Rapport de calcul
Carte Horaire
Compte rendu de calcul Bernese GPS Software
 Informations sur le processus
Informations sur le processus Sessions intégrées au calcul
Sessions intégrées au calcul Orbites et coordonnées du pôle
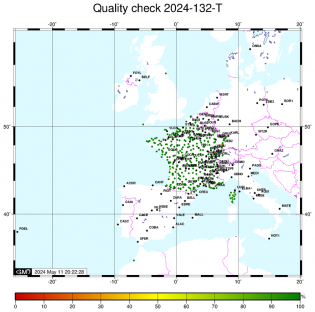
Orbites et coordonnées du pôle Controle qualité
Controle qualité Calcul sur le code
Calcul sur le code Fixation des ambiguités
Fixation des ambiguités Fixation des ambiguités Core
Fixation des ambiguités Core Estimation des ZTD finaux (coordonnées fixées) - ADDNEQ2
Estimation des ZTD finaux (coordonnées fixées) - ADDNEQ2



Sessions intégrées au calcul
| Année | DOY | Session |
|---|---|---|
| 2026 | 069 | p |
| 2026 | 069 | q |
| 2026 | 069 | r |
| 2026 | 069 | s |
| 2026 | 069 | t |
| 2026 | 069 | u |




Orbites et coordonnées du pôle
| Orbites | Coordonnées du pôle |
|---|---|
| IGS0OPSULT_20260681800_02D_15M_ORB.SP3.gz | IGS0OPSULT_20260681800_02D_01D_ERP.ERP.gz |




Controle qualité
| Station | Dernier fichier | Ratio des fichiers précédents | Ratio qc_gps | Ratio qc_glo | qc - header GPS | qc - header GLO | IOD slips <10° | IOD slips >10° | Station utilisée dans le calcul |
|---|---|---|---|---|---|---|---|---|---|
| AJAC | - | 0.0% | - | - | - | - | - | - | - |
| ALPE | - | 100.0% | - | - | - | - | - | - | - |
| BANN | - | 100.0% | - | - | - | - | - | - | - |
| BIAZ | - | 0.0% | - | - | - | - | - | - | - |
| BRET | - | 0.0% | - | - | - | - | - | - | - |
| BVOI | - | 100.0% | - | - | - | - | - | - | - |
| BVS2 | - | 100.0% | - | - | - | - | - | - | - |
| CABW | - | 0.0% | - | - | - | - | - | - | - |
| CAMO | - | 0.0% | - | - | - | - | - | - | - |
| CANE | - | 0.0% | - | - | - | - | - | - | - |
| CAUS | - | 0.0% | - | - | - | - | - | - | - |
| CH2T | - | 100.0% | - | - | - | - | - | - | - |
| CHBL | - | 100.0% | - | - | - | - | - | - | - |
| CHE2 | - | 100.0% | - | - | - | - | - | - | - |
| CHIZ | - | 0.0% | - | - | - | - | - | - | - |
| CHOL | - | 100.0% | - | - | - | - | - | - | - |
| CHPO | - | 100.0% | - | - | - | - | - | - | - |
| CHTL | - | 0.0% | - | - | - | - | - | - | - |
| COCR | - | 100.0% | - | - | - | - | - | - | - |
| CONN | - | 100.0% | - | - | - | - | - | - | - |
| CORZ | - | 100.0% | - | - | - | - | - | - | - |
| CRAU | - | 100.0% | - | - | - | - | - | - | - |
| CRTS | - | 100.0% | - | - | - | - | - | - | - |
| CSTO | - | 100.0% | - | - | - | - | - | - | - |
| CULA | - | 100.0% | - | - | - | - | - | - | - |
| CYLM | - | 100.0% | - | - | - | - | - | - | - |
| DBMH | - | 100.0% | - | - | - | - | - | - | - |
| DENT | - | 0.0% | - | - | - | - | - | - | - |
| DEQU | - | 100.0% | - | - | - | - | - | - | - |
| DHUI | - | 100.0% | - | - | - | - | - | - | - |
| DOCX | - | 100.0% | - | - | - | - | - | - | - |
| DOMP | - | 100.0% | - | - | - | - | - | - | - |
| DRUL | - | 100.0% | - | - | - | - | - | - | - |
| EBN2 | - | 100.0% | - | - | - | - | - | - | - |
| EQH2 | - | 100.0% | - | - | - | - | - | - | - |
| FEUR | - | 100.0% | - | - | - | - | - | - | - |
| FJC2 | - | 0.0% | - | - | - | - | - | - | - |
| GAJN | - | 100.0% | - | - | - | - | - | - | - |
| GINA | - | 0.0% | - | - | - | - | - | - | - |
| ILBO | - | 0.0% | - | - | - | - | - | - | - |
| JANU | - | 100.0% | - | - | - | - | - | - | - |
| JOUS | - | 100.0% | - | - | - | - | - | - | - |
| KEGA | - | 100.0% | - | - | - | - | - | - | - |
| LAIG | - | 100.0% | - | - | - | - | - | - | - |
| LAVL | - | 100.0% | - | - | - | - | - | - | - |
| LBRD | - | 0.0% | - | - | - | - | - | - | - |
| LBUG | - | 0.0% | - | - | - | - | - | - | - |
| LCST | - | 100.0% | - | - | - | - | - | - | - |
| LCVA | - | 100.0% | - | - | - | - | - | - | - |
| LESA | - | 100.0% | - | - | - | - | - | - | - |
| LEZI | - | 100.0% | - | - | - | - | - | - | - |
| LIZ2 | - | 100.0% | - | - | - | - | - | - | - |
| LLGV | - | 100.0% | - | - | - | - | - | - | - |
| LMDM | - | 100.0% | - | - | - | - | - | - | - |
| LOYT | - | 100.0% | - | - | - | - | - | - | - |
| MAGR | - | 100.0% | - | - | - | - | - | - | - |
| MAKS | - | 0.0% | - | - | - | - | - | - | - |
| MATC | - | 100.0% | - | - | - | - | - | - | - |
| MAUP | - | 100.0% | - | - | - | - | - | - | - |
| MTP2 | - | 0.0% | - | - | - | - | - | - | - |
| NGER | - | 100.0% | - | - | - | - | - | - | - |
| NMTR | - | 100.0% | - | - | - | - | - | - | - |
| NOGT | - | 100.0% | - | - | - | - | - | - | - |
| NOYL | - | 100.0% | - | - | - | - | - | - | - |
| NVPT | - | 100.0% | - | - | - | - | - | - | - |
| OMBA | - | 100.0% | - | - | - | - | - | - | - |
| PALI | - | 0.0% | - | - | - | - | - | - | - |
| PAMO | - | 100.0% | - | - | - | - | - | - | - |
| PAYE | - | 0.0% | - | - | - | - | - | - | - |
| PEY2 | - | 100.0% | - | - | - | - | - | - | - |
| PLGN | - | 100.0% | - | - | - | - | - | - | - |
| PLMG | - | 100.0% | - | - | - | - | - | - | - |
| PLST | - | 100.0% | - | - | - | - | - | - | - |
| PNTV | - | 100.0% | - | - | - | - | - | - | - |
| POBU | - | 0.0% | - | - | - | - | - | - | - |
| PUYA | - | 100.0% | - | - | - | - | - | - | - |
| ROSD | - | 100.0% | - | - | - | - | - | - | - |
| ROSI | - | 0.0% | - | - | - | - | - | - | - |
| SCOA | - | 0.0% | - | - | - | - | - | - | - |
| SCOP | - | 0.0% | - | - | - | - | - | - | - |
| SEM2 | - | 100.0% | - | - | - | - | - | - | - |
| SJDV | - | 0.0% | - | - | - | - | - | - | - |
| SMCR | - | 100.0% | - | - | - | - | - | - | - |
| STMR | - | 0.0% | - | - | - | - | - | - | - |
| VEN3 | - | 100.0% | - | - | - | - | - | - | - |
| VIN1 | - | 100.0% | - | - | - | - | - | - | - |
| VLPL | - | 100.0% | - | - | - | - | - | - | - |
| VNTE | - | 100.0% | - | - | - | - | - | - | - |
| VOUZ | - | 100.0% | - | - | - | - | - | - | - |
| VRCT | - | 100.0% | - | - | - | - | - | - | - |
| VSOU | - | 100.0% | - | - | - | - | - | - | - |
| WARE | - | 0.0% | - | - | - | - | - | - | - |
| ZIMM | - | 0.0% | - | - | - | - | - | - | - |
| MODA | x | 100.0% | 81.6% | - | 3.9380m | - | 7 | 20 | x |
| OSAN | x | 100.0% | 83.8% | - | 7.5070m | - | 130 | 12 | x |
| VILD | x | 100.0% | 84.7% | - | 7.1100m | - | 1 | 6 | x |
| RMZT | x | 100.0% | 85% | - | 5.2275m | - | 0 | 3 | x |
| FILF | x | 100.0% | 86.1% | - | 9.8283m | - | 17 | 4 | x |
| FLRC | x | 100.0% | 86.2% | - | 6.1122m | - | 27 | 5 | x |
| MAUL | x | 100.0% | 87.7% | - | 7.0263m | - | 258 | 34 | x |
| FIED | x | 100.0% | 87.9% | - | 7.2251m | - | 61 | 68 | x |
| LAHE | x | 100.0% | 87.9% | - | 12.6742m | - | 274 | 69 | x |
| PIAA | x | 100.0% | 88.5% | - | 5.3697m | - | 34 | 10 | x |
| ELBA | x | 100.0% | 89.2% | - | 7.4646m | - | 38 | 22 | x |
| CDBC | x | 100.0% | 89.6% | - | 10.0782m | - | 54 | 19 | x |
| STJ9 | x | 100.0% | 89.8% | - | 10.7699m | - | 76 | 31 | x |
| UNME | x | 100.0% | 90.1% | - | 11.8800m | - | 157 | 2 | x |
| CRTE | x | 100.0% | 90.3% | - | 4.7740m | - | 6 | 12 | x |
| HOLA | x | 100.0% | 90.7% | - | 10.2386m | - | 159 | 1 | x |
| MSGT | x | 100.0% | 90.8% | - | 7.5593m | - | 3 | 2 | x |
| GENO | x | 100.0% | 91.7% | - | 7.5448m | - | 142 | 40 | x |
| CHTG | x | 100.0% | 91.9% | - | 11.8016m | - | 47 | 12 | x |
| TUDA | x | 100.0% | 91.9% | - | 7.6081m | - | 84 | 6 | x |
| CROL | x | 100.0% | 92.4% | - | 6.3212m | - | 48 | 0 | x |
| ANDE | x | 100.0% | 92.9% | - | 7.8140m | - | 0 | 0 | x |
| BRDO | x | 100.0% | 93.1% | - | 8.1405m | - | 43 | 3 | x |
| ARUF | x | 100.0% | 93.3% | - | 8.0294m | - | 77 | 10 | x |
| CAMA | x | 100.0% | 93.3% | - | 8.4904m | - | 198 | 11 | x |
| RIXH | x | 100.0% | 93.3% | - | 7.6353m | - | 16 | 9 | x |
| SAFF | x | 100.0% | 93.3% | - | 7.0148m | - | 121 | 0 | x |
| LACA | x | 100.0% | 93.6% | - | 8.0176m | - | 263 | 19 | x |
| LUCE | x | 100.0% | 93.9% | - | 11.8467m | - | 49 | 7 | x |
| MARS | x | 100.0% | 94.2% | - | 8.6584m | - | 240 | 13 | x |
| SARZ | x | 100.0% | 94.2% | - | 9.1123m | - | 24 | 13 | x |
| BACT | x | 100.0% | 94.8% | - | 7.0121m | - | 0 | 1 | x |
| MARG | x | 100.0% | 94.9% | - | 6.2988m | - | 4 | 4 | x |
| MDEU | x | 100.0% | 94.9% | - | 16.1062m | - | 23 | 3 | x |
| MSMM | x | 100.0% | 94.9% | - | 8.0948m | - | 182 | 7 | x |
| SGIL | x | 100.0% | 95% | - | 8.4597m | - | 153 | 46 | x |
| DIPP | x | 100.0% | 95.3% | - | 11.1730m | - | 55 | 14 | x |
| TORI | x | 100.0% | 95.3% | - | 8.2062m | - | 57 | 35 | x |
| MELN | x | 100.0% | 95.4% | - | 9.9284m | - | 24 | 4 | x |
| PUYV | x | 100.0% | 95.6% | - | 8.8576m | - | 57 | 7 | x |
| STNZ | x | 100.0% | 95.6% | - | 7.3712m | - | 13 | 3 | x |
| BOUS | x | 100.0% | 95.8% | - | 6.8138m | - | 8 | 1 | x |
| BUSI | x | 100.0% | 95.9% | - | 6.9388m | - | 0 | 1 | x |
| DOCO | x | 100.0% | 95.9% | - | 8.5025m | - | 119 | 47 | x |
| VIRG | x | 100.0% | 95.9% | - | 10.4140m | - | 199 | 17 | x |
| BLG2 | x | 100.0% | 96% | - | 7.9629m | - | 7 | 3 | x |
| BSCN | x | 100.0% | 96% | - | 6.0720m | - | 117 | 36 | x |
| MLLC | x | 100.0% | 96.1% | - | 6.4108m | - | 3 | 6 | x |
| MRON | x | 100.0% | 96.1% | - | 9.8696m | - | 32 | 4 | x |
| COVI | x | 100.0% | 96.2% | - | 8.8189m | - | 46 | 18 | x |
| DOUL | x | 100.0% | 96.2% | - | 9.8337m | - | 2 | 1 | x |
| NIVO | x | 100.0% | 96.2% | - | 7.4329m | - | 0 | 4 | x |
| PRR2 | x | 100.0% | 96.2% | - | 6.6804m | - | 0 | 2 | x |
| SOLR | x | 100.0% | 96.2% | - | 5.9410m | - | 77 | 7 | x |
| LLIV | x | 100.0% | 96.3% | - | 7.8690m | - | 211 | 71 | x |
| LUCI | x | 100.0% | 96.3% | - | 7.6913m | - | 0 | 3 | x |
| ERCK | x | 100.0% | 96.4% | - | 6.9236m | - | 14 | 1 | x |
| PRAC | x | 100.0% | 96.4% | - | 7.1803m | - | 98 | 31 | x |
| BEAV | x | 100.0% | 96.5% | - | 7.3864m | - | 7 | 1 | x |
| FRTT | x | 100.0% | 96.5% | - | 8.7852m | - | 60 | 3 | x |
| STMA | x | 100.0% | 96.5% | - | 7.4551m | - | 0 | 0 | x |
| OISO | x | 100.0% | 96.6% | - | 9.5189m | - | 90 | 1 | x |
| SJPL | x | 100.0% | 96.6% | - | 7.8970m | - | 137 | 6 | x |
| THNV | x | 100.0% | 96.6% | - | 8.3467m | - | 0 | 0 | x |
| VDOM | x | 100.0% | 96.6% | - | 9.2929m | - | 50 | 1 | x |
| CEVY | x | 100.0% | 96.7% | - | 10.4125m | - | 28 | 15 | x |
| AGDS | x | 100.0% | 96.8% | - | 7.9364m | - | 0 | 4 | x |
| COAU | x | 100.0% | 96.8% | - | 8.6386m | - | 26 | 3 | x |
| EZEV | x | 100.0% | 96.8% | - | 8.6500m | - | 149 | 4 | x |
| MICH | x | 100.0% | 96.8% | - | 7.5871m | - | 78 | 6 | x |
| SAFQ | x | 100.0% | 96.8% | - | 8.2447m | - | 0 | 0 | x |
| LURI | x | 100.0% | 96.9% | - | 9.7905m | - | 0 | 3 | x |
| PAYR | x | 100.0% | 96.9% | - | 9.6533m | - | 180 | 3 | x |
| SCDA | x | 100.0% | 96.9% | - | 7.5323m | - | 64 | 1 | x |
| UNIX | x | 100.0% | 96.9% | - | 8.6151m | - | 55 | 2 | x |
| AICI | x | 100.0% | 97% | - | 8.9661m | - | 0 | 1 | x |
| BAUG | x | 100.0% | 97% | - | 8.4264m | - | 0 | 0 | x |
| PTRC | x | 100.0% | 97% | - | 6.7724m | - | 0 | 3 | x |
| PUY2 | x | 100.0% | 97% | - | 7.5115m | - | 156 | 1 | x |
| ROMY | x | 100.0% | 97% | - | 7.2523m | - | 0 | 3 | x |
| SARS | x | 100.0% | 97% | - | 6.5873m | - | 71 | 18 | x |
| SRMG | x | 100.0% | 97% | - | 7.8748m | - | 151 | 3 | x |
| STS2 | x | 100.0% | 97% | - | 8.5966m | - | 0 | 2 | x |
| ZARA | x | 100.0% | 97% | - | 12.3635m | - | 203 | 41 | x |
| AGN2 | x | 100.0% | 97.1% | - | 9.4570m | - | 0 | 0 | x |
| ATH2 | x | 100.0% | 97.1% | - | 9.1494m | - | 0 | 0 | x |
| BMNT | x | 100.0% | 97.1% | - | 9.2993m | - | 206 | 11 | x |
| CANT | x | 100.0% | 97.1% | - | 12.5535m | - | 58 | 1 | x |
| CAPT | x | 100.0% | 97.1% | - | 8.3763m | - | 0 | 0 | x |
| CHDY | x | 100.0% | 97.1% | - | 8.3434m | - | 15 | 0 | x |
| DUNQ | x | 100.0% | 97.1% | - | 12.4224m | - | 66 | 4 | x |
| HERB | x | 100.0% | 97.1% | - | 9.2541m | - | 0 | 4 | x |
| NMCU | x | 100.0% | 97.1% | - | 6.8239m | - | 15 | 1 | x |
| RAYL | x | 100.0% | 97.1% | - | 8.6469m | - | 147 | 4 | x |
| RIO1 | x | 100.0% | 97.1% | - | 7.9359m | - | 205 | 17 | x |
| SBL2 | x | 100.0% | 97.1% | - | 7.7750m | - | 24 | 0 | x |
| SIST | x | 100.0% | 97.1% | - | 4.9979m | - | 0 | 4 | x |
| STGS | x | 100.0% | 97.1% | - | 7.8277m | - | 0 | 6 | x |
| ANGL | x | 100.0% | 97.2% | - | 7.5590m | - | 41 | 1 | x |
| BRST | x | 100.0% | 97.2% | - | 11.3286m | - | 81 | 1 | x |
| C2CN | x | 100.0% | 97.2% | - | 5.8641m | - | 0 | 0 | x |
| CRAL | x | 100.0% | 97.2% | - | 6.5828m | - | 185 | 13 | x |
| IRAF | x | 100.0% | 97.2% | - | 5.5985m | - | 192 | 9 | x |
| LPPZ | x | 100.0% | 97.2% | - | 7.8273m | - | 39 | 0 | x |
| LUMI | x | 100.0% | 97.2% | - | 9.2250m | - | 99 | 6 | x |
| MGIS | x | 100.0% | 97.2% | - | 8.5826m | - | 62 | 19 | x |
| RNNE | x | 100.0% | 97.2% | - | 9.8980m | - | 24 | 13 | x |
| ROTG | x | 100.0% | 97.2% | - | 11.5451m | - | 73 | 5 | x |
| RUPT | x | 100.0% | 97.2% | - | 5.7780m | - | 0 | 1 | x |
| SAMR | x | 100.0% | 97.2% | - | 8.0375m | - | 0 | 2 | x |
| SARL | x | 100.0% | 97.2% | - | 9.5701m | - | 30 | 3 | x |
| TANZ | x | 100.0% | 97.2% | - | 9.3776m | - | 31 | 6 | x |
| AMNS | x | 100.0% | 97.3% | - | 8.1055m | - | 0 | 3 | x |
| APT2 | x | 100.0% | 97.3% | - | 7.1203m | - | 0 | 0 | x |
| ATST | x | 100.0% | 97.3% | - | 7.6429m | - | 239 | 11 | x |
| AUCH | x | 100.0% | 97.3% | - | 9.1206m | - | 63 | 1 | x |
| BARD | x | 100.0% | 97.3% | - | 6.2890m | - | 0 | 1 | x |
| BLVR | x | 100.0% | 97.3% | - | 7.5918m | - | 126 | 3 | x |
| BMHG | x | 100.0% | 97.3% | - | 8.6653m | - | 76 | 4 | x |
| BRIV | x | 100.0% | 97.3% | - | 8.7138m | - | 132 | 14 | x |
| CAMP | x | 100.0% | 97.3% | - | 9.1579m | - | 148 | 16 | x |
| CAUD | x | 100.0% | 97.3% | - | 9.2216m | - | 5 | 0 | x |
| CERN | x | 100.0% | 97.3% | - | 9.1734m | - | 55 | 5 | x |
| CHBR | x | 100.0% | 97.3% | - | 8.8933m | - | 21 | 1 | x |
| DOUA | x | 100.0% | 97.3% | - | 9.6127m | - | 11 | 1 | x |
| FAJP | x | 100.0% | 97.3% | - | 9.2986m | - | 237 | 13 | x |
| GUIP | x | 100.0% | 97.3% | - | 7.1891m | - | 73 | 0 | x |
| HRSN | x | 100.0% | 97.3% | - | 8.9530m | - | 7 | 0 | x |
| LACO | x | 100.0% | 97.3% | - | 7.9226m | - | 241 | 9 | x |
| LCAN | x | 100.0% | 97.3% | - | 8.6872m | - | 128 | 15 | x |
| LGAR | x | 100.0% | 97.3% | - | 8.8001m | - | 144 | 2 | x |
| LIL2 | x | 100.0% | 97.3% | - | 6.4120m | - | 36 | 0 | x |
| MNBL | x | 100.0% | 97.3% | - | 6.4606m | - | 0 | 0 | x |
| MTDM | x | 100.0% | 97.3% | - | 7.9510m | - | 175 | 0 | x |
| OTER | x | 100.0% | 97.3% | - | 7.7816m | - | 42 | 3 | x |
| QNSN | x | 100.0% | 97.3% | - | 7.1998m | - | 0 | 1 | x |
| TGRI | x | 100.0% | 97.3% | - | 8.9284m | - | 0 | 1 | x |
| THIL | x | 100.0% | 97.3% | - | 9.7089m | - | 2 | 0 | x |
| TRMO | x | 100.0% | 97.3% | - | 8.2734m | - | 151 | 6 | x |
| AGLM | x | 100.0% | 97.4% | - | 8.6007m | - | 115 | 14 | x |
| ARDN | x | 100.0% | 97.4% | - | 7.6586m | - | 0 | 2 | x |
| AVRA | x | 100.0% | 97.4% | - | 7.6754m | - | 11 | 1 | x |
| BRE2 | x | 100.0% | 97.4% | - | 6.1903m | - | 0 | 0 | x |
| BRM2 | x | 100.0% | 97.4% | - | 7.8446m | - | 0 | 3 | x |
| CHIO | x | 100.0% | 97.4% | - | 6365350.4781m | - | 47 | 0 | x |
| CHTA | x | 100.0% | 97.4% | - | 8.0038m | - | 47 | 3 | x |
| ETOI | x | 100.0% | 97.4% | - | 10.2649m | - | 63 | 15 | x |
| FAYE | x | 100.0% | 97.4% | - | 7.0901m | - | 92 | 5 | x |
| GDIJ | x | 100.0% | 97.4% | - | 5.4862m | - | 0 | 0 | x |
| HERS | x | 100.0% | 97.4% | - | 9.7964m | - | 36 | 0 | x |
| ISLA | x | 100.0% | 97.4% | - | 9.9600m | - | 42 | 0 | x |
| LGES | x | 100.0% | 97.4% | - | 9.2068m | - | 0 | 0 | x |
| LONS | x | 100.0% | 97.4% | - | 10.5481m | - | 50 | 5 | x |
| MAZE | x | 100.0% | 97.4% | - | 8.3677m | - | 0 | 0 | x |
| MOFA | x | 100.0% | 97.4% | - | 6.8449m | - | 0 | 2 | x |
| MOMO | x | 100.0% | 97.4% | - | 8.3407m | - | 0 | 0 | x |
| MTIL | x | 100.0% | 97.4% | - | 7.5809m | - | 0 | 0 | x |
| PMTH | x | 100.0% | 97.4% | - | 6365662.6842m | - | 24 | 0 | x |
| QUYC | x | 100.0% | 97.4% | - | 7.5760m | - | 0 | 0 | x |
| SNNT | x | 100.0% | 97.4% | - | 5.4066m | - | 0 | 7 | x |
| STV2 | x | 100.0% | 97.4% | - | 7.8968m | - | 6 | 2 | x |
| UCAG | x | 100.0% | 97.4% | - | 9.9139m | - | 250 | 127 | x |
| VISN | x | 100.0% | 97.4% | - | 8.7652m | - | 183 | 2 | x |
| ALGY | x | 100.0% | 97.5% | - | 4.6488m | - | 0 | 0 | x |
| AMB2 | x | 100.0% | 97.5% | - | 8.6970m | - | 99 | 8 | x |
| ASNE | x | 100.0% | 97.5% | - | 7.7557m | - | 0 | 0 | x |
| AUNI | x | 100.0% | 97.5% | - | 9.3774m | - | 0 | 0 | x |
| BAL2 | x | 100.0% | 97.5% | - | 7.8319m | - | 0 | 0 | x |
| BEUG | x | 100.0% | 97.5% | - | 8.1814m | - | 0 | 0 | x |
| BOGU | x | 100.0% | 97.5% | - | 8.7441m | - | 24 | 1 | x |
| BRG9 | x | 100.0% | 97.5% | - | 6.5084m | - | 0 | 0 | x |
| COUT | x | 100.0% | 97.5% | - | 8.6813m | - | 185 | 3 | x |
| CRHX | x | 100.0% | 97.5% | - | 8.1333m | - | 54 | 0 | x |
| CUBX | x | 100.0% | 97.5% | - | 9.6803m | - | 101 | 1 | x |
| DGLG | x | 100.0% | 97.5% | - | 10.9907m | - | 117 | 25 | x |
| DOJX | x | 100.0% | 97.5% | - | 9.2040m | - | 29 | 1 | x |
| EGLT | x | 100.0% | 97.5% | - | 8.1260m | - | 78 | 4 | x |
| FLER | x | 100.0% | 97.5% | - | 7.8350m | - | 16 | 2 | x |
| FOUG | x | 100.0% | 97.5% | - | 8.9900m | - | 45 | 3 | x |
| GIE1 | x | 100.0% | 97.5% | - | 6.9259m | - | 0 | 1 | x |
| GORN | x | 100.0% | 97.5% | - | 9.3047m | - | 54 | 2 | x |
| JARG | x | 100.0% | 97.5% | - | 9.4956m | - | 41 | 2 | x |
| JOI2 | x | 100.0% | 97.5% | - | 8.2244m | - | 47 | 1 | x |
| KONE | x | 100.0% | 97.5% | - | 7.9150m | - | 42 | 0 | x |
| LROC | x | 100.0% | 97.5% | - | 11.3093m | - | 81 | 9 | x |
| MIRE | x | 100.0% | 97.5% | - | 8.3826m | - | 0 | 0 | x |
| MPRV | x | 100.0% | 97.5% | - | 6.8160m | - | 0 | 0 | x |
| MTMN | x | 100.0% | 97.5% | - | 9.2618m | - | 91 | 1 | x |
| MULH | x | 100.0% | 97.5% | - | 8.7892m | - | 10 | 0 | x |
| NARB | x | 100.0% | 97.5% | - | 9.6694m | - | 146 | 26 | x |
| RENN | x | 100.0% | 97.5% | - | 8.7337m | - | 23 | 0 | x |
| RYO2 | x | 100.0% | 97.5% | - | 6.4446m | - | 0 | 0 | x |
| SAUN | x | 100.0% | 97.5% | - | 6.7013m | - | 0 | 0 | x |
| SMLE | x | 100.0% | 97.5% | - | 8.9190m | - | 106 | 1 | x |
| SPIO | x | 100.0% | 97.5% | - | 7.4576m | - | 0 | 0 | x |
| STGN | x | 100.0% | 97.5% | - | 7.2896m | - | 0 | 5 | x |
| TLSE | x | 100.0% | 97.5% | - | 10.9268m | - | 224 | 3 | x |
| TRBL | x | 100.0% | 97.5% | - | 8.0808m | - | 0 | 1 | x |
| UZER | x | 100.0% | 97.5% | - | 9.3704m | - | 0 | 1 | x |
| VAUD | x | 100.0% | 97.5% | - | 5.9631m | - | 0 | 0 | x |
| VFCH | x | 100.0% | 97.5% | - | 7.1498m | - | 70 | 2 | x |
| AIGL | x | 100.0% | 97.6% | - | 9.3693m | - | 273 | 0 | x |
| ANGE | x | 100.0% | 97.6% | - | 9.7808m | - | 30 | 0 | x |
| ANNY | x | 100.0% | 97.6% | - | 8.6680m | - | 7 | 1 | x |
| ARCI | x | 100.0% | 97.6% | - | 9.0272m | - | 4 | 1 | x |
| AXPV | x | 100.0% | 97.6% | - | 6.7624m | - | 139 | 3 | x |
| BREV | x | 100.0% | 97.6% | - | 8.7801m | - | 22 | 0 | x |
| BRMZ | x | 100.0% | 97.6% | - | 8.9695m | - | 45 | 2 | x |
| CALS | x | 100.0% | 97.6% | - | 9.8131m | - | 74 | 2 | x |
| CASE | x | 100.0% | 97.6% | - | 9.6042m | - | 266 | 38 | x |
| CBRY | x | 100.0% | 97.6% | - | 8.1785m | - | 9 | 5 | x |
| CEPI | x | 100.0% | 97.6% | - | 9.6564m | - | 188 | 15 | x |
| CHAS | x | 100.0% | 97.6% | - | 9.2105m | - | 38 | 1 | x |
| CHBS | x | 100.0% | 97.6% | - | 8.7153m | - | 70 | 0 | x |
| CHLN | x | 100.0% | 97.6% | - | 9.5716m | - | 43 | 1 | x |
| CHRE | x | 100.0% | 97.6% | - | 8.5130m | - | 3 | 0 | x |
| CHRX | x | 100.0% | 97.6% | - | 8.7192m | - | 161 | 0 | x |
| CNTL | x | 100.0% | 97.6% | - | 10.4081m | - | 79 | 1 | x |
| CRSM | x | 100.0% | 97.6% | - | 9.2743m | - | 31 | 0 | x |
| CRVA | x | 100.0% | 97.6% | - | 10.3738m | - | 3 | 0 | x |
| DAGO | x | 100.0% | 97.6% | - | 9.9513m | - | 86 | 2 | x |
| DIPL | x | 100.0% | 97.6% | - | 8.0633m | - | 33 | 0 | x |
| DRUS | x | 100.0% | 97.6% | - | 9.0935m | - | 33 | 0 | x |
| ENTZ | x | 100.0% | 97.6% | - | 7.2159m | - | 137 | 0 | x |
| EPR2 | x | 100.0% | 97.6% | - | 8.3760m | - | 50 | 0 | x |
| ESCO | x | 100.0% | 97.6% | - | 6.6998m | - | 63 | 21 | x |
| EUSK | x | 100.0% | 97.6% | - | 9.4053m | - | 222 | 0 | x |
| FCMP | x | 100.0% | 97.6% | - | 8.3718m | - | 21 | 3 | x |
| FDET | x | 100.0% | 97.6% | - | 10.0895m | - | 174 | 8 | x |
| FEGA | x | 100.0% | 97.6% | - | 9.7231m | - | 67 | 0 | x |
| FET2 | x | 100.0% | 97.6% | - | 10.6639m | - | 14 | 0 | x |
| FOU2 | x | 100.0% | 97.6% | - | 9.0124m | - | 45 | 0 | x |
| GARD | x | 100.0% | 97.6% | - | 8.3115m | - | 0 | 0 | x |
| GLRA | x | 100.0% | 97.6% | - | 8.4732m | - | 104 | 0 | x |
| GRAC | x | 100.0% | 97.6% | - | 7.4923m | - | 244 | 1 | x |
| GRAS | x | 100.0% | 97.6% | - | 8.4587m | - | 256 | 2 | x |
| GRON | x | 100.0% | 97.6% | - | 9.8925m | - | 95 | 2 | x |
| HONF | x | 100.0% | 97.6% | - | 10.3070m | - | 72 | 1 | x |
| HOUL | x | 100.0% | 97.6% | - | 10.1194m | - | 114 | 0 | x |
| IGNP | x | 100.0% | 97.6% | - | 12.4087m | - | 70 | 0 | x |
| ILDX | x | 100.0% | 97.6% | - | 7.6390m | - | 142 | 0 | x |
| JONZ | x | 100.0% | 97.6% | - | 9.3539m | - | 145 | 0 | x |
| KARL | x | 100.0% | 97.6% | - | 11.7781m | - | 58 | 0 | x |
| LENE | x | 100.0% | 97.6% | - | 8.9049m | - | 30 | 1 | x |
| LETO | x | 100.0% | 97.6% | - | 9.1631m | - | 29 | 0 | x |
| LGBO | x | 100.0% | 97.6% | - | 9.0750m | - | 187 | 0 | x |
| LPAS | x | 100.0% | 97.6% | - | 8.2225m | - | 26 | 0 | x |
| MACH | x | 100.0% | 97.6% | - | 9.4863m | - | 101 | 0 | x |
| MAN2 | x | 100.0% | 97.6% | - | 10.1436m | - | 45 | 0 | x |
| MLVL | x | 100.0% | 97.6% | - | 8.6056m | - | 78 | 0 | x |
| MONS | x | 100.0% | 97.6% | - | 6.2782m | - | 0 | 2 | x |
| MTBT | x | 100.0% | 97.6% | - | 9.6111m | - | 27 | 2 | x |
| NACY | x | 100.0% | 97.6% | - | 11.8292m | - | 42 | 0 | x |
| NAYC | x | 100.0% | 97.6% | - | 13.3512m | - | 193 | 2 | x |
| NICA | x | 100.0% | 97.6% | - | 9.6784m | - | 0 | 4 | x |
| PERP | x | 100.0% | 97.6% | - | 9.6496m | - | 109 | 18 | x |
| PESS | x | 100.0% | 97.6% | - | 9.0663m | - | 192 | 2 | x |
| PLEM | x | 100.0% | 97.6% | - | 8.0604m | - | 40 | 0 | x |
| PRNY | x | 100.0% | 97.6% | - | 9.9103m | - | 68 | 7 | x |
| PZNA | x | 100.0% | 97.6% | - | 10.2071m | - | 247 | 5 | x |
| REDO | x | 100.0% | 97.6% | - | 8.9535m | - | 52 | 0 | x |
| RGNC | x | 100.0% | 97.6% | - | 6.7619m | - | 0 | 1 | x |
| ROYA | x | 100.0% | 97.6% | - | 8.4587m | - | 143 | 0 | x |
| RST2 | x | 100.0% | 97.6% | - | 9.4996m | - | 242 | 1 | x |
| SEES | x | 100.0% | 97.6% | - | 9.1603m | - | 22 | 1 | x |
| SET1 | x | 100.0% | 97.6% | - | 10.3109m | - | 293 | 8 | x |
| SEUR | x | 100.0% | 97.6% | - | 8.1886m | - | 53 | 2 | x |
| SIRT | x | 100.0% | 97.6% | - | 9.2461m | - | 0 | 0 | x |
| SJDA | x | 100.0% | 97.6% | - | 9.8882m | - | 183 | 9 | x |
| SMSP | x | 100.0% | 97.6% | - | 9.7417m | - | 22 | 0 | x |
| SOUS | x | 100.0% | 97.6% | - | 7.5241m | - | 143 | 0 | x |
| STBR | x | 100.0% | 97.6% | - | 9.4663m | - | 3 | 0 | x |
| STBX | x | 100.0% | 97.6% | - | 10.9723m | - | 89 | 8 | x |
| STJF | x | 100.0% | 97.6% | - | 10.5829m | - | 88 | 1 | x |
| STLO | x | 100.0% | 97.6% | - | 8.1574m | - | 7 | 0 | x |
| TLMF | x | 100.0% | 97.6% | - | 6.9621m | - | 250 | 3 | x |
| TLSG | x | 100.0% | 97.6% | - | 9.3126m | - | 196 | 3 | x |
| TLTG | x | 100.0% | 97.6% | - | 13.7482m | - | 205 | 6 | x |
| TRYS | x | 100.0% | 97.6% | - | 7.5804m | - | 0 | 4 | x |
| VERN | x | 100.0% | 97.6% | - | 7.1298m | - | 0 | 3 | x |
| YPOR | x | 100.0% | 97.6% | - | 8.2357m | - | 55 | 0 | x |
| ALU2 | x | 100.0% | 97.7% | - | 7.2679m | - | 0 | 0 | x |
| CACI | x | 100.0% | 97.7% | - | 8.7031m | - | 62 | 2 | x |
| OPME | x | 100.0% | 97.7% | - | 5.2671m | - | 0 | 1 | x |
| STAZ | x | 100.0% | 97.7% | - | 5.7267m | - | 0 | 0 | x |
| STF2 | x | 100.0% | 97.7% | - | 6.9834m | - | 0 | 4 | x |
| AVAL | x | 100.0% | 97.8% | - | 8.0039m | - | 36 | 5 | x |
| CLFD | x | 100.0% | 97.8% | - | 4.4294m | - | 0 | 1 | x |
| LULI | x | 100.0% | 97.8% | - | 10.2266m | - | 154 | 10 | x |
| AILT | x | 100.0% | 97.9% | - | 9.8332m | - | 89 | 0 | x |
| ANAY | x | 100.0% | 97.9% | - | 9.4025m | - | 128 | 15 | x |
| AUTN | x | 100.0% | 97.9% | - | 8.5541m | - | 12 | 1 | x |
| BRMF | x | 100.0% | 97.9% | - | 8.7371m | - | 44 | 1 | x |
| CLFE | x | 100.0% | 97.9% | - | 7.8889m | - | 117 | 0 | x |
| DROU | x | 100.0% | 97.9% | - | 9.1231m | - | 100 | 1 | x |
| STPS | x | 100.0% | 97.9% | - | 9.7082m | - | 121 | 0 | x |
| VOUR | x | 100.0% | 97.9% | - | 11.8648m | - | 110 | 1 | x |




Calcul des coordonnées sur le code
Les stations sont éliminées du calcul si l'écart entre les coordonnées issues du calcul sur le code et les coordonnées initiales est supérieur à dE=10.0 m, dN=10.0 m ou dU=20.0 m| Station | dE (m) | dN (m) | dU (m) |
|---|---|---|---|
| AGDS | 0.01m | 0.18m | 0.63m |
| AGLM | 0.00m | -0.13m | -0.28m |
| AGN2 | -0.06m | -0.05m | 0.33m |
| AICI | -0.02m | 0.24m | 0.38m |
| AIGL | 0.17m | -0.10m | 0.55m |
| AILT | 0.04m | 0.12m | 0.44m |
| ALGY | 0.05m | 0.23m | 0.13m |
| ALU2 | 0.04m | 0.23m | 0.51m |
| AMB2 | -0.09m | -0.11m | -0.27m |
| AMNS | -0.06m | 0.23m | -0.04m |
| ANAY | 0.10m | 0.02m | 0.56m |
| ANDE | 0.10m | 0.07m | 0.37m |
| ANGE | 0.08m | 0.16m | 0.33m |
| ANGL | 0.11m | 0.34m | 0.24m |
| ANNY | 0.10m | -0.04m | 0.37m |
| APT2 | 0.02m | 0.29m | -0.13m |
| ARCI | 0.12m | 0.09m | 0.21m |
| ARDN | -0.14m | 0.08m | -0.06m |
| ARUF | 0.22m | 0.40m | 0.49m |
| ASNE | 0.03m | 0.19m | 0.41m |
| ATH2 | -0.01m | 0.24m | 0.67m |
| ATST | -0.08m | 0.03m | 0.01m |
| AUCH | 0.34m | 0.19m | 0.41m |
| AUNI | 0.10m | 0.31m | 0.66m |
| AUTN | -0.06m | 0.28m | 0.04m |
| AVAL | 0.17m | 0.36m | 0.53m |
| AVRA | 0.02m | 0.14m | 0.11m |
| AXPV | -0.26m | 0.67m | 0.69m |
| BACT | 0.07m | 0.09m | 0.00m |
| BAL2 | -0.09m | 0.19m | 0.76m |
| BARD | 0.02m | 0.15m | 0.41m |
| BAUG | -0.07m | 0.17m | 0.43m |
| BEAV | 0.31m | 0.32m | 0.41m |
| BEUG | 0.02m | 0.20m | 0.59m |
| BLG2 | -0.09m | 0.17m | 0.12m |
| BLVR | 0.60m | 0.35m | 1.12m |
| BMHG | 0.26m | 0.00m | 0.07m |
| BMNT | 0.25m | 0.23m | 0.44m |
| BOGU | 0.10m | -0.15m | -0.23m |
| BOUS | 0.04m | 0.12m | 0.41m |
| BRDO | 0.04m | 0.36m | 0.52m |
| BRE2 | -0.12m | 0.33m | 0.60m |
| BREV | 0.12m | 0.31m | 0.30m |
| BRG9 | 0.02m | 0.31m | 0.18m |
| BRIV | 0.11m | 0.21m | 0.81m |
| BRM2 | 0.02m | 0.21m | 0.44m |
| BRMF | -0.03m | 0.18m | 0.11m |
| BRMZ | 0.01m | 0.25m | 0.38m |
| BRST | -0.14m | 0.16m | 1.02m |
| BSCN | -0.01m | 0.40m | 0.04m |
| BUSI | 0.03m | 0.15m | 0.58m |
| C2CN | -0.00m | 0.32m | 0.41m |
| CACI | 0.18m | -0.08m | 0.40m |
| CALS | 0.19m | 0.12m | 0.46m |
| CAMA | 0.19m | 0.36m | 0.62m |
| CAMP | -0.01m | 0.24m | 0.63m |
| CANT | -0.08m | 0.24m | 0.63m |
| CAPT | -0.22m | 0.14m | 0.20m |
| CASE | -0.21m | 0.28m | 0.38m |
| CAUD | 0.14m | 0.18m | 0.14m |
| CBRY | 0.15m | 0.15m | 0.56m |
| CDBC | 0.02m | -0.04m | 0.18m |
| CEPI | 0.18m | 0.22m | 0.52m |
| CERN | 0.11m | 0.36m | 0.53m |
| CEVY | 0.27m | 0.16m | 0.68m |
| CHAS | 0.22m | 0.13m | 0.47m |
| CHBR | 0.08m | -0.15m | -0.01m |
| CHBS | 0.24m | 0.11m | 0.38m |
| CHDY | 0.08m | 0.09m | 0.24m |
| CHIO | 0.17m | 0.05m | 0.77m |
| CHLN | 0.23m | -0.16m | -0.23m |
| CHRE | 0.20m | -0.03m | 0.73m |
| CHRX | 0.12m | -0.00m | 0.36m |
| CHTA | 0.18m | 0.12m | 0.16m |
| CHTG | 0.05m | 0.29m | 1.38m |
| CLFD | -0.06m | 0.24m | 0.77m |
| CLFE | 0.17m | 0.18m | 0.61m |
| CNTL | 0.16m | 0.01m | 0.24m |
| COAU | 0.12m | 0.06m | 0.22m |
| COUT | 0.21m | 0.23m | 0.23m |
| COVI | 0.18m | 0.01m | 0.37m |
| CRAL | 0.24m | 0.08m | 0.53m |
| CRHX | 0.01m | 0.15m | -0.01m |
| CROL | -0.06m | -0.00m | 0.14m |
| CRSM | 0.09m | 0.18m | 0.20m |
| CRTE | 0.06m | 0.43m | 0.31m |
| CRVA | 0.12m | 0.06m | 0.49m |
| CUBX | -0.09m | -0.60m | -0.38m |
| DAGO | 0.16m | 0.03m | 0.15m |
| DGLG | 0.13m | 0.14m | 0.30m |
| DIPL | 0.31m | 0.04m | 0.33m |
| DIPP | 0.06m | 0.13m | 0.09m |
| DOCO | 0.19m | 0.46m | 0.27m |
| DOJX | 0.01m | 0.09m | 0.10m |
| DOUA | 0.16m | 0.10m | 0.48m |
| DOUL | 0.08m | 0.14m | 0.36m |
| DROU | 0.02m | 0.11m | 0.22m |
| DRUS | 0.11m | 0.07m | 0.63m |
| DUNQ | 0.07m | 0.00m | 0.36m |
| EGLT | -0.06m | 0.36m | -0.02m |
| ELBA | 0.10m | 0.51m | 0.37m |
| ENTZ | 0.04m | 0.27m | 0.24m |
| EPR2 | 0.22m | 0.18m | 0.21m |
| ERCK | 0.16m | 0.07m | 1.17m |
| ESCO | -0.06m | 0.12m | -0.08m |
| ETOI | 0.01m | 0.10m | 0.33m |
| EUSK | -0.09m | -0.06m | 1.03m |
| EZEV | -0.09m | -0.00m | -0.06m |
| FAJP | 0.38m | 0.09m | 0.62m |
| FAYE | 0.33m | 0.33m | 0.58m |
| FCMP | 0.10m | 0.09m | 0.42m |
| FDET | 0.17m | 0.07m | 0.53m |
| FEGA | 0.10m | 0.10m | 0.46m |
| FET2 | 0.09m | 0.34m | 0.34m |
| FIED | 0.28m | 0.09m | 1.28m |
| FILF | 0.12m | -0.23m | 0.78m |
| FLER | 0.16m | -0.04m | -0.17m |
| FLRC | -0.15m | 0.22m | 0.59m |
| FOU2 | 0.23m | 0.08m | 0.22m |
| FOUG | 0.15m | 0.10m | 0.34m |
| FRTT | 0.07m | 0.27m | 0.36m |
| GARD | -0.18m | -0.04m | 0.51m |
| GDIJ | -0.04m | 0.30m | 0.09m |
| GENO | -0.16m | 0.34m | 0.20m |
| GIE1 | 0.01m | 0.14m | 0.60m |
| GLRA | 0.03m | 0.11m | 0.92m |
| GORN | -0.10m | 0.12m | -0.13m |
| GRAC | -0.14m | 0.40m | 0.05m |
| GRAS | 0.32m | 0.31m | 0.56m |
| GRON | 0.15m | 0.16m | 0.44m |
| GUIP | -0.03m | 0.33m | 0.09m |
| HERB | -0.06m | 0.30m | 0.59m |
| HERS | -0.11m | 0.05m | 0.53m |
| HOLA | 0.09m | 0.26m | 0.55m |
| HONF | 0.05m | 0.07m | 0.41m |
| HOUL | 0.14m | 0.04m | 0.37m |
| HRSN | 0.20m | 0.14m | 0.34m |
| IGNP | -0.18m | -0.06m | 0.39m |
| ILDX | -0.04m | 0.36m | 0.28m |
| IRAF | 0.13m | 0.17m | 0.36m |
| ISLA | 0.10m | 0.13m | 0.66m |
| JARG | 0.15m | -0.02m | -0.08m |
| JOI2 | -0.15m | -0.15m | -0.37m |
| JONZ | 0.23m | 0.12m | 0.39m |
| KARL | 0.14m | 0.07m | 0.89m |
| KONE | 0.06m | 0.06m | 0.00m |
| LACA | -0.03m | 0.47m | 1.28m |
| LACO | 0.01m | 0.18m | 0.43m |
| LAHE | -0.36m | -0.04m | 0.39m |
| LCAN | 0.36m | 0.23m | 0.72m |
| LENE | 0.11m | 0.13m | -0.09m |
| LETO | 0.31m | 0.14m | 0.17m |
| LGAR | -0.07m | -0.13m | -0.31m |
| LGBO | 0.08m | -0.04m | 0.11m |
| LGES | 0.04m | 0.22m | 0.64m |
| LIL2 | 0.01m | 0.16m | -0.06m |
| LLIV | -0.38m | 0.08m | 0.33m |
| LONS | 0.18m | -0.02m | 0.56m |
| LPAS | 0.05m | 0.13m | 0.10m |
| LPPZ | 0.20m | -0.09m | 0.59m |
| LROC | -0.07m | -0.01m | 0.33m |
| LUCE | 0.11m | 0.26m | 1.43m |
| LUCI | -0.02m | 0.27m | -0.07m |
| LULI | 0.02m | -0.04m | 0.53m |
| LUMI | 0.02m | 0.21m | 0.39m |
| LURI | 0.15m | 0.27m | 1.10m |
| MACH | 0.20m | -0.04m | 0.30m |
| MAN2 | 0.04m | 0.26m | 0.47m |
| MARG | -0.01m | 0.17m | -0.05m |
| MARS | -0.18m | 0.07m | -0.05m |
| MAUL | 0.08m | -0.07m | 0.46m |
| MAZE | 0.02m | 0.20m | 0.63m |
| MDEU | 0.03m | 0.26m | -0.04m |
| MELN | 0.11m | 0.28m | 0.47m |
| MGIS | 0.58m | 0.56m | 0.60m |
| MICH | 0.19m | 0.22m | 0.63m |
| MIRE | 0.04m | 0.14m | 0.47m |
| MLLC | 0.12m | -0.15m | 0.26m |
| MLVL | 0.02m | 0.23m | 0.00m |
| MNBL | 0.05m | 0.14m | 0.09m |
| MODA | 0.25m | 0.16m | -0.17m |
| MOFA | -0.07m | 0.19m | 0.45m |
| MOMO | 0.06m | 0.29m | 0.65m |
| MONS | -0.55m | 0.46m | 1.22m |
| MPRV | -0.09m | 0.29m | 0.68m |
| MRON | 0.14m | 0.12m | 0.43m |
| MSGT | 0.22m | 0.09m | 0.38m |
| MSMM | -0.15m | -0.08m | -0.06m |
| MTBT | 0.02m | 0.12m | 0.09m |
| MTDM | 0.15m | 0.10m | 0.68m |
| MTIL | -0.04m | -0.04m | -0.26m |
| MTMN | 0.22m | 0.13m | -0.25m |
| MULH | 0.23m | 0.23m | 0.67m |
| NACY | -0.02m | 0.19m | 0.68m |
| NARB | 0.20m | 0.31m | 0.32m |
| NAYC | 0.16m | -0.04m | 0.46m |
| NICA | 0.06m | 0.09m | 0.12m |
| NIVO | 0.07m | 0.22m | 0.64m |
| NMCU | 0.25m | -0.65m | -0.07m |
| OISO | -0.26m | 0.14m | -0.13m |
| OPME | 0.15m | 0.35m | 0.48m |
| OSAN | 0.05m | 0.55m | 0.18m |
| OTER | -0.01m | 0.26m | -0.25m |
| PAYR | -0.78m | 0.52m | 0.52m |
| PERP | 0.09m | 0.20m | 0.38m |
| PESS | 0.09m | 0.16m | 0.35m |
| PIAA | 0.17m | -0.31m | 1.02m |
| PLEM | 0.11m | 0.20m | 0.47m |
| PMTH | 0.11m | 0.10m | 0.72m |
| PRAC | 0.10m | -0.15m | 0.67m |
| PRNY | 0.16m | 0.07m | 0.66m |
| PRR2 | 0.02m | 0.36m | 0.40m |
| PTRC | 0.05m | 0.05m | 0.31m |
| PUY2 | -0.12m | -0.18m | -0.43m |
| PUYV | 0.00m | 0.59m | 0.35m |
| PZNA | 0.20m | 0.13m | 0.62m |
| QNSN | -0.15m | 0.38m | 0.76m |
| QUYC | -0.09m | 0.16m | 0.22m |
| RAYL | -0.17m | -0.21m | -0.25m |
| REDO | 0.17m | 0.15m | 0.38m |
| RENN | 0.10m | 0.13m | 0.33m |
| RGNC | 0.14m | 0.24m | 0.42m |
| RIO1 | -0.18m | 0.25m | 0.54m |
| RIXH | 0.21m | 0.17m | 0.65m |
| RMZT | -0.15m | 0.22m | 0.59m |
| RNNE | 0.23m | 0.11m | 0.86m |
| ROMY | -0.06m | 0.28m | 0.61m |
| ROTG | 0.01m | 0.21m | 0.63m |
| ROYA | 0.13m | 0.29m | 0.44m |
| RST2 | 0.24m | 0.37m | 0.72m |
| RUPT | 0.02m | 0.19m | 0.62m |
| RYO2 | -0.17m | 0.14m | 0.68m |
| SAFF | 0.04m | 0.08m | 0.57m |
| SAFQ | 0.04m | 0.14m | 0.57m |
| SAMR | -0.04m | 0.02m | 0.39m |
| SARL | 0.11m | 0.12m | 0.21m |
| SARS | 0.14m | 0.36m | 0.49m |
| SARZ | 0.07m | -0.44m | 0.09m |
| SAUN | -0.12m | 0.19m | 0.31m |
| SBL2 | -0.22m | 0.02m | 0.36m |
| SCDA | 0.08m | 0.08m | 0.37m |
| SEES | 0.19m | 0.15m | -0.08m |
| SET1 | 0.04m | 0.26m | 0.91m |
| SEUR | 0.03m | -0.19m | -0.10m |
| SGIL | 0.18m | 0.05m | 0.24m |
| SIRT | 0.06m | 0.45m | 0.44m |
| SIST | 0.03m | 0.33m | 0.49m |
| SJDA | 0.26m | 0.10m | 0.47m |
| SJPL | 0.36m | 0.39m | 0.42m |
| SMLE | 0.25m | 0.04m | -0.28m |
| SMSP | 0.08m | 0.19m | 0.54m |
| SNNT | -0.01m | 0.13m | 0.75m |
| SOLR | 0.20m | 0.14m | -0.20m |
| SOUS | 0.06m | 0.12m | 0.40m |
| SPIO | -0.17m | 0.13m | 0.25m |
| SRMG | 0.07m | -0.03m | 0.10m |
| STAZ | -0.07m | 0.14m | 0.38m |
| STBR | 0.01m | 0.28m | 0.15m |
| STBX | 0.26m | 0.26m | 0.74m |
| STF2 | 0.02m | 0.16m | 0.67m |
| STGN | 0.11m | 0.30m | 0.61m |
| STGS | -0.11m | 0.09m | -0.24m |
| STJ9 | 0.23m | 0.34m | 1.66m |
| STJF | 0.03m | -0.05m | 0.56m |
| STLO | 0.28m | 0.29m | 0.08m |
| STMA | 0.12m | 0.29m | 0.31m |
| STNZ | 0.80m | -0.57m | 1.02m |
| STPS | 0.06m | 0.11m | 0.46m |
| STS2 | 0.05m | 0.33m | 0.12m |
| STV2 | 0.05m | 0.12m | 0.21m |
| TANZ | 0.10m | 0.14m | 0.19m |
| TGRI | -0.40m | 0.66m | 1.01m |
| THIL | 0.11m | 0.15m | 0.52m |
| THNV | -0.03m | 0.02m | 0.28m |
| TLMF | -0.21m | 0.18m | 0.07m |
| TLSE | -0.16m | -0.02m | 1.11m |
| TLSG | -0.19m | 0.33m | 0.61m |
| TLTG | -0.02m | -0.03m | 0.35m |
| TORI | -0.18m | 0.42m | 0.77m |
| TRBL | -0.02m | 0.21m | 0.64m |
| TRMO | -0.09m | -0.15m | -0.19m |
| TRYS | -0.02m | 0.14m | 0.74m |
| TUDA | 0.10m | 0.19m | 0.77m |
| UCAG | -0.11m | 0.16m | 0.59m |
| UNIX | 0.04m | -0.02m | -0.19m |
| UNME | 0.16m | 0.23m | 0.91m |
| UZER | 0.12m | 0.14m | 0.25m |
| VAUD | 0.11m | 0.15m | 0.75m |
| VDOM | -0.08m | 0.25m | 0.46m |
| VERN | -0.04m | 0.26m | 0.42m |
| VFCH | -0.08m | 0.42m | 0.31m |
| VILD | 0.00m | 0.22m | 0.32m |
| VIRG | -0.01m | 0.20m | 0.24m |
| VISN | 0.07m | 0.14m | 0.73m |
| VOUR | 0.30m | 0.20m | 0.54m |
| YPOR | -0.06m | -0.03m | 0.24m |
| ZARA | 0.01m | 0.32m | 0.90m |




Fixation des ambiguités
| Station 1 | Station 2 | Longueur | Constellation | Ambiguités posées | Ambiguités fixées QIF | Ambiguités fixées L12 | Ratio |
|---|---|---|---|---|---|---|---|
| GRAC | GRAS | 0.032km | G | 40 | 34 | 5 | 97.500% |
| OSAN | PIAA | 9.910km | G | 40 | 26 | 10 | 90.000% |
| APT2 | RST2 | 10.538km | G | 42 | 38 | 4 | 100.000% |
| BRDO | LURI | 12.642km | G | 36 | 34 | 0 | 94.444% |
| MSMM | QNSN | 16.958km | G | 44 | 34 | 9 | 97.727% |
| MICH | RST2 | 18.862km | G | 48 | 36 | 10 | 95.833% |
| ATST | CRTE | 20.958km | G | 38 | 30 | - | 78.9% |
| BRDO | TUDA | 21.050km | G | 34 | 32 | - | 94.1% |
| EZEV | NICA | 23.146km | G | 52 | 30 | - | 57.7% |
| FAYE | GRAC | 23.864km | G | 36 | 36 | - | 100.0% |
| GRAC | NICA | 25.367km | G | 46 | 32 | - | 69.6% |
| LUCI | TUDA | 27.929km | G | 52 | 40 | - | 76.9% |
| ATST | LUCI | 29.073km | G | 52 | 42 | - | 80.8% |
| CAMA | RAYL | 29.272km | G | 38 | 34 | - | 89.5% |
| MICH | QNSN | 33.591km | G | 54 | 36 | - | 66.7% |
| ATST | SARS | 34.652km | G | 40 | 34 | - | 85.0% |
| LUMI | OSAN | 34.874km | G | 34 | 26 | - | 76.5% |
| MICH | SIST | 37.330km | G | 78 | 38 | - | 48.7% |
| CAMP | PIAA | 37.642km | G | 62 | 24 | - | 38.7% |
| CAMA | FAYE | 38.152km | G | 44 | 36 | - | 81.8% |
| SIST | STV2 | 40.585km | G | 64 | 28 | - | 43.8% |
| CRTE | OSAN | 43.003km | G | 36 | 30 | - | 83.3% |
| APT2 | AXPV | 43.016km | G | 42 | 36 | - | 85.7% |
| CAMA | QNSN | 43.800km | G | 46 | 38 | - | 82.6% |
| CAMA | TLTG | 46.042km | G | 44 | 34 | - | 77.3% |
| BACT | STV2 | 47.604km | G | 40 | 40 | - | 100.0% |
| CAMP | MAUL | 48.537km | G | 42 | 30 | - | 71.4% |
| SIST | RMZT | 48.822km | G | 50 | 30 | - | 60.0% |
| BRDO | ELBA | 60.313km | G | 32 | 28 | - | 87.5% |
| MAUL | MDEU | 70.520km | G | 38 | 28 | - | 73.7% |
| ELBA | VIRG | 100.349km | G | 40 | 30 | - | 75.0% |
| BACT | TORI | 109.842km | G | 44 | 34 | - | 77.3% |
| GENO | TORI | 122.792km | G | 46 | 34 | - | 73.9% |
| GENO | VIRG | 154.028km | G | 42 | 32 | - | 76.2% |
| MDEU | UCAG | 184.570km | G | 36 | 28 | - | 77.8% |
| AXPV | MARS | 23.663km | G | 38 | 30 | - | 78.9% |
| TLSE | TLSG | 1.266km | G | 44 | 32 | 12 | 100.000% |
| SAFF | SAFQ | 2.717km | G | 38 | 34 | 4 | 100.000% |
| AGDS | PZNA | 15.801km | G | 38 | 32 | 4 | 94.737% |
| AGDS | SET1 | 20.829km | G | 46 | 38 | - | 82.6% |
| CEPI | FAJP | 21.558km | G | 40 | 32 | - | 80.0% |
| AIGL | FLRC | 22.705km | G | 36 | 32 | - | 88.9% |
| AGN2 | LGAR | 23.213km | G | 48 | 32 | - | 66.7% |
| HOLA | SAFF | 26.190km | G | 38 | 28 | - | 73.7% |
| CRAL | STGS | 26.785km | G | 60 | 40 | - | 66.7% |
| LACA | SAFQ | 32.780km | G | 58 | 34 | - | 58.6% |
| AIGL | HOLA | 33.408km | G | 50 | 36 | - | 72.0% |
| LACA | PAYR | 34.474km | G | 56 | 36 | - | 64.3% |
| FILF | LLIV | 37.619km | G | 50 | 34 | - | 68.0% |
| SAFQ | TRMO | 37.889km | G | 40 | 34 | - | 85.0% |
| LACO | STGS | 38.675km | G | 48 | 36 | - | 75.0% |
| FILF | PERP | 40.759km | G | 40 | 34 | - | 85.0% |
| AUCH | BMNT | 40.760km | G | 58 | 32 | - | 55.2% |
| AGDS | NARB | 42.678km | G | 46 | 34 | - | 73.9% |
| LACO | TLSG | 43.906km | G | 34 | 34 | - | 100.0% |
| AGN2 | BMNT | 43.972km | G | 54 | 36 | - | 66.7% |
| FAJP | PAYR | 45.918km | G | 42 | 36 | - | 85.7% |
| MTIL | PRAC | 48.200km | G | 48 | 32 | - | 66.7% |
| FLRC | STBX | 48.638km | G | 36 | 30 | - | 83.3% |
| FAJP | MSGT | 48.679km | G | 38 | 34 | - | 89.5% |
| ESCO | STGS | 51.626km | G | 48 | 36 | - | 75.0% |
| LLIV | MSGT | 52.764km | G | 42 | 30 | - | 71.4% |
| BMNT | TLSE | 53.716km | G | 50 | 38 | - | 76.0% |
| BMNT | MTIL | 55.833km | G | 56 | 38 | - | 67.9% |
| NARB | PERP | 57.039km | G | 38 | 34 | - | 89.5% |
| ESCO | MSGT | 57.393km | G | 40 | 32 | - | 80.0% |
| FLRC | SCDA | 58.268km | G | 36 | 34 | - | 94.4% |
| SET1 | SGIL | 66.696km | G | 40 | 34 | - | 85.0% |
| PRAC | SOUS | 77.929km | G | 44 | 34 | - | 77.3% |
| CASE | FILF | 85.324km | G | 40 | 32 | - | 80.0% |
| ESCO | ZARA | 193.529km | G | 38 | 34 | - | 89.5% |
| TLMF | TLSE | 8.686km | G | 52 | 34 | 17 | 98.077% |
| CUBX | PESS | 7.665km | G | 40 | 34 | 6 | 100.000% |
| CHRX | MPRV | 10.035km | G | 48 | 22 | 23 | 93.750% |
| MTDM | STS2 | 13.562km | G | 38 | 32 | 6 | 100.000% |
| AICI | MLLC | 16.545km | G | 52 | 38 | 9 | 90.385% |
| JONZ | MAZE | 18.428km | G | 42 | 36 | 3 | 92.857% |
| C2CN | SRMG | 19.565km | G | 42 | 34 | 7 | 97.619% |
| GARD | SRMG | 21.513km | G | 46 | 38 | - | 82.6% |
| AUNI | LROC | 21.794km | G | 38 | 34 | - | 89.5% |
| AICI | PUY2 | 22.996km | G | 58 | 28 | - | 48.3% |
| LROC | SPIO | 23.360km | G | 48 | 34 | - | 70.8% |
| IRAF | MLLC | 24.856km | G | 44 | 30 | - | 68.2% |
| ARUF | UNME | 26.119km | G | 42 | 34 | - | 81.0% |
| BRIV | UZER | 29.404km | G | 62 | 40 | - | 64.5% |
| BARD | COUT | 30.252km | G | 48 | 36 | - | 75.0% |
| ANGL | LROC | 30.965km | G | 44 | 32 | - | 72.7% |
| QUYC | ROYA | 31.769km | G | 44 | 32 | - | 72.7% |
| C2CN | LGAR | 33.888km | G | 38 | 34 | - | 89.5% |
| MAZE | ROYA | 34.513km | G | 46 | 34 | - | 73.9% |
| BARD | JONZ | 37.522km | G | 46 | 34 | - | 73.9% |
| AUNI | SJDA | 38.131km | G | 48 | 32 | - | 66.7% |
| ARUF | MLLC | 38.787km | G | 42 | 34 | - | 81.0% |
| PUY2 | STS2 | 39.326km | G | 52 | 32 | - | 61.5% |
| AGLM | BARD | 39.994km | G | 50 | 36 | - | 72.0% |
| COUT | CUBX | 40.253km | G | 46 | 32 | - | 69.6% |
| COUT | GARD | 40.684km | G | 46 | 34 | - | 73.9% |
| LCAN | PESS | 41.684km | G | 46 | 34 | - | 73.9% |
| ROYA | SPIO | 42.358km | G | 60 | 34 | - | 56.7% |
| ANGL | CHTA | 42.746km | G | 46 | 28 | - | 60.9% |
| AGLM | LGBO | 45.459km | G | 36 | 34 | - | 94.4% |
| CAPT | MTDM | 49.341km | G | 42 | 34 | - | 81.0% |
| CAPT | LGAR | 50.721km | G | 38 | 36 | - | 94.7% |
| SJDA | SMLE | 55.975km | G | 50 | 32 | - | 64.0% |
| AGLM | CHRX | 57.570km | G | 50 | 34 | - | 68.0% |
| BRIV | LGBO | 75.120km | G | 48 | 40 | - | 83.3% |
| IRAF | RIO1 | 130.144km | G | 52 | 32 | - | 61.5% |
| CANT | RIO1 | 158.306km | G | 54 | 42 | - | 77.8% |
| ILDX | SPIO | 12.169km | G | 48 | 36 | 12 | 100.000% |
| CHBS | VFCH | 6.813km | G | 34 | 32 | 2 | 100.000% |
| ARDN | JARG | 20.796km | G | 40 | 34 | - | 85.0% |
| BOUS | SNNT | 23.295km | G | 58 | 40 | - | 69.0% |
| ALU2 | CACI | 23.317km | G | 40 | 30 | - | 75.0% |
| BRMZ | MAN2 | 24.118km | G | 36 | 34 | - | 94.4% |
| SAUN | VDOM | 26.238km | G | 38 | 34 | - | 89.5% |
| MONS | STPS | 26.700km | G | 42 | 32 | - | 76.2% |
| FDET | PTRC | 28.160km | G | 56 | 30 | - | 53.6% |
| FDET | RGNC | 30.122km | G | 52 | 38 | - | 73.1% |
| FDET | SAUN | 30.296km | G | 44 | 36 | - | 81.8% |
| LULI | MONS | 32.467km | G | 48 | 32 | - | 66.7% |
| JARG | OISO | 32.628km | G | 52 | 46 | - | 88.5% |
| ASNE | GIE1 | 32.876km | G | 46 | 32 | - | 69.6% |
| OTER | RGNC | 35.116km | G | 52 | 38 | - | 73.1% |
| MPRV | MTMN | 39.471km | G | 50 | 24 | - | 48.0% |
| GIE1 | MGIS | 40.658km | G | 40 | 32 | - | 80.0% |
| BAUG | PTRC | 41.645km | G | 48 | 30 | - | 62.5% |
| BRG9 | LGES | 43.739km | G | 46 | 36 | - | 78.3% |
| ARCI | TRBL | 45.321km | G | 38 | 34 | - | 89.5% |
| ARCI | BRMZ | 45.582km | G | 40 | 34 | - | 85.0% |
| JARG | MGIS | 46.102km | G | 52 | 32 | - | 61.5% |
| GRON | LULI | 46.301km | G | 48 | 32 | - | 66.7% |
| MTMN | NAYC | 47.493km | G | 38 | 32 | - | 84.2% |
| CHBS | STMA | 47.614km | G | 56 | 44 | - | 78.6% |
| ASNE | GRON | 47.709km | G | 40 | 30 | - | 75.0% |
| OTER | STMA | 48.202km | G | 56 | 40 | - | 71.4% |
| AVAL | CACI | 48.613km | G | 48 | 36 | - | 75.0% |
| BRMZ | VDOM | 48.702km | G | 40 | 32 | - | 80.0% |
| AILT | MGIS | 49.226km | G | 36 | 32 | - | 88.9% |
| BOUS | BRG9 | 52.009km | G | 38 | 34 | - | 89.5% |
| NAYC | OTER | 54.539km | G | 50 | 34 | - | 68.0% |
| ASNE | VFCH | 54.793km | G | 44 | 32 | - | 72.7% |
| AILT | AVAL | 59.814km | G | 46 | 32 | - | 69.6% |
| MELN | MGIS | 62.754km | G | 42 | 32 | - | 76.2% |
| BRG9 | EGLT | 66.011km | G | 36 | 32 | - | 88.9% |
| LGES | MPRV | 67.373km | G | 52 | 18 | - | 34.6% |
| BRE2 | NAYC | 68.362km | G | 70 | 36 | - | 51.4% |
| REDO | SAMR | 5.304km | G | 66 | 32 | 30 | 93.939% |
| BRST | GUIP | 9.498km | G | 52 | 36 | 16 | 100.000% |
| CRSM | EPR2 | 15.171km | G | 40 | 32 | 8 | 100.000% |
| BRST | LPPZ | 20.853km | G | 50 | 36 | - | 72.0% |
| HERB | STNZ | 21.046km | G | 44 | 34 | - | 77.3% |
| EPR2 | HOUL | 24.167km | G | 44 | 34 | - | 77.3% |
| HOUL | LAHE | 24.992km | G | 54 | 26 | - | 48.1% |
| PRR2 | STLO | 27.609km | G | 66 | 36 | - | 54.5% |
| HERB | REDO | 28.467km | G | 48 | 32 | - | 66.7% |
| RYO2 | CHTA | 29.431km | G | 54 | 36 | - | 66.7% |
| MACH | NMCU | 30.954km | G | 40 | 34 | - | 85.0% |
| HERB | SARZ | 34.992km | G | 60 | 26 | - | 43.3% |
| LPAS | SARZ | 36.676km | G | 54 | 26 | - | 48.1% |
| PLEM | STBR | 39.079km | G | 50 | 32 | - | 64.0% |
| FLER | GORN | 41.694km | G | 42 | 32 | - | 76.2% |
| MACH | RYO2 | 42.636km | G | 48 | 38 | - | 79.2% |
| CHBR | COVI | 43.233km | G | 44 | 34 | - | 77.3% |
| MACH | STNZ | 44.497km | G | 42 | 36 | - | 85.7% |
| ANGE | SBL2 | 44.560km | G | 38 | 34 | - | 89.5% |
| COVI | SBL2 | 44.716km | G | 46 | 34 | - | 73.9% |
| GUIP | ROTG | 44.840km | G | 54 | 32 | - | 59.3% |
| CHBR | RENN | 48.522km | G | 40 | 32 | - | 80.0% |
| CRSM | STLO | 49.617km | G | 42 | 34 | - | 81.0% |
| CHBR | SAMR | 50.123km | G | 68 | 32 | - | 47.1% |
| AVRA | GORN | 50.759km | G | 44 | 34 | - | 77.3% |
| DIPL | STBR | 50.859km | G | 48 | 26 | - | 54.2% |
| JOI2 | MACH | 51.073km | G | 40 | 32 | - | 80.0% |
| CRHX | KONE | 52.269km | G | 44 | 30 | - | 68.2% |
| AVRA | STLO | 53.437km | G | 44 | 30 | - | 68.2% |
| AVRA | DIPL | 53.532km | G | 40 | 34 | - | 85.0% |
| COVI | GORN | 53.639km | G | 46 | 34 | - | 73.9% |
| FLER | SEES | 55.780km | G | 44 | 34 | - | 77.3% |
| CRHX | ROTG | 57.270km | G | 54 | 40 | - | 74.1% |
| BMHG | PRR2 | 60.185km | G | 64 | 28 | - | 43.8% |
| CRHX | STBR | 65.263km | G | 54 | 32 | - | 59.3% |
| BMHG | CHIO | 168.038km | G | 44 | 34 | - | 77.3% |
| BMHG | PMTH | 184.821km | G | 58 | 38 | - | 65.5% |
| BMHG | CHTG | 14.047km | G | 58 | 40 | 16 | 96.552% |
| FCMP | YPOR | 4.899km | G | 38 | 34 | 2 | 94.737% |
| DGLG | DUNQ | 6.246km | G | 44 | 34 | 10 | 100.000% |
| BOGU | CNTL | 6.737km | G | 42 | 36 | 4 | 95.238% |
| BUSI | CAUD | 11.213km | G | 48 | 32 | 16 | 100.000% |
| IGNP | MLVL | 11.918km | G | 36 | 32 | 4 | 100.000% |
| ATH2 | COAU | 13.352km | G | 44 | 36 | 4 | 90.909% |
| HONF | LAHE | 14.986km | G | 64 | 28 | 33 | 95.312% |
| CDBC | STJF | 15.516km | G | 44 | 34 | 9 | 97.727% |
| CDBC | MOFA | 16.842km | G | 54 | 34 | 16 | 92.593% |
| FCMP | MOFA | 18.192km | G | 58 | 34 | 22 | 96.552% |
| HONF | STJF | 21.154km | G | 46 | 34 | - | 73.9% |
| IGNP | SIRT | 21.675km | G | 40 | 32 | - | 80.0% |
| SIRT | TGRI | 23.887km | G | 42 | 36 | - | 85.7% |
| DOUA | LIL2 | 24.378km | G | 50 | 26 | - | 52.0% |
| CDBC | CNTL | 24.616km | G | 42 | 32 | - | 76.2% |
| FOU2 | MOMO | 28.158km | G | 40 | 36 | - | 90.0% |
| BOGU | MOMO | 29.212km | G | 44 | 36 | - | 81.8% |
| IGNP | ISLA | 32.986km | G | 36 | 32 | - | 88.9% |
| BREV | TGRI | 33.056km | G | 38 | 34 | - | 89.5% |
| BEUG | DOUA | 33.457km | G | 44 | 32 | - | 72.7% |
| CNTL | LENE | 34.268km | G | 46 | 38 | - | 82.6% |
| AMNS | DOUL | 34.704km | G | 46 | 36 | - | 78.3% |
| CALS | DGLG | 35.346km | G | 50 | 34 | - | 68.0% |
| BEUG | CHLN | 36.076km | G | 36 | 34 | - | 94.4% |
| CAUD | DOUA | 37.082km | G | 42 | 32 | - | 76.2% |
| BEUG | DOUL | 37.680km | G | 38 | 32 | - | 84.2% |
| BEAV | ISLA | 38.808km | G | 42 | 32 | - | 76.2% |
| BUSI | COAU | 43.552km | G | 50 | 40 | - | 80.0% |
| ATH2 | FET2 | 43.849km | G | 40 | 34 | - | 85.0% |
| CRVA | FET2 | 43.946km | G | 36 | 32 | - | 88.9% |
| BUSI | HRSN | 45.012km | G | 50 | 40 | - | 80.0% |
| AMNS | BEAV | 46.939km | G | 50 | 36 | - | 72.0% |
| AMNS | FOU2 | 47.333km | G | 40 | 34 | - | 85.0% |
| FEGA | FET2 | 47.962km | G | 38 | 34 | - | 89.5% |
| CALS | LETO | 52.605km | G | 44 | 36 | - | 81.8% |
| DOUL | LETO | 65.226km | G | 36 | 32 | - | 88.9% |
| HERS | LETO | 98.805km | G | 38 | 34 | - | 89.5% |
| DIPP | FOU2 | 36.962km | G | 42 | 34 | - | 81.0% |
| MULH | RIXH | 4.343km | G | 44 | 38 | 6 | 100.000% |
| DOJX | RUPT | 7.482km | G | 64 | 32 | 27 | 92.188% |
| ENTZ | STJ9 | 8.669km | G | 48 | 30 | 16 | 95.833% |
| BRM2 | STJ9 | 10.101km | G | 60 | 34 | 23 | 95.000% |
| CHRE | MIRE | 14.628km | G | 40 | 34 | 5 | 97.500% |
| ROMY | THIL | 16.340km | G | 44 | 36 | 7 | 97.727% |
| BRM2 | DRUS | 19.007km | G | 50 | 36 | 8 | 88.000% |
| ROMY | FET2 | 19.149km | G | 44 | 36 | 8 | 100.000% |
| BLVR | MNBL | 24.159km | G | 52 | 32 | - | 61.5% |
| ALGY | FRTT | 24.554km | G | 50 | 36 | - | 72.0% |
| BAL2 | RIXH | 25.113km | G | 46 | 34 | - | 73.9% |
| ERCK | SARL | 28.326km | G | 42 | 36 | - | 85.7% |
| DOCO | THNV | 30.225km | G | 50 | 34 | - | 68.0% |
| BRM2 | ERCK | 30.931km | G | 50 | 34 | - | 68.0% |
| ANDE | DAGO | 32.200km | G | 40 | 30 | - | 75.0% |
| LUCE | MULH | 33.713km | G | 58 | 32 | - | 55.2% |
| ENTZ | TANZ | 35.277km | G | 44 | 34 | - | 77.3% |
| LUCE | MNBL | 35.421km | G | 54 | 28 | - | 51.9% |
| BAL2 | TANZ | 36.488km | G | 46 | 38 | - | 82.6% |
| ALGY | BSCN | 37.489km | G | 54 | 38 | - | 70.4% |
| CHRE | NACY | 38.102km | G | 34 | 34 | - | 100.0% |
| ANDE | RUPT | 42.514km | G | 58 | 26 | - | 44.8% |
| MTBT | RUPT | 43.031km | G | 66 | 30 | - | 45.5% |
| DRUS | KARL | 43.317km | G | 42 | 38 | - | 90.5% |
| ALGY | GDIJ | 44.736km | G | 58 | 38 | - | 65.5% |
| ANDE | SMSP | 45.251km | G | 36 | 28 | - | 77.8% |
| MTBT | TRYS | 46.345km | G | 54 | 30 | - | 55.6% |
| BLVR | BSCN | 47.816km | G | 50 | 36 | - | 72.0% |
| SMSP | THIL | 48.319km | G | 34 | 30 | - | 88.2% |
| FOUG | MIRE | 49.137km | G | 38 | 34 | - | 89.5% |
| CEVY | DOCO | 49.942km | G | 54 | 32 | - | 59.3% |
| DOCO | NACY | 51.086km | G | 46 | 32 | - | 69.6% |
| FOUG | MNBL | 54.413km | G | 44 | 32 | - | 72.7% |
| CEVY | SMSP | 54.898km | G | 44 | 32 | - | 72.7% |
| CEVY | CHDY | 60.540km | G | 44 | 36 | - | 81.8% |
| CHAS | TRYS | 62.991km | G | 58 | 36 | - | 62.1% |
| EUSK | THNV | 151.811km | G | 34 | 34 | - | 100.0% |
| ETOI | STJ9 | 7.383km | G | 64 | 36 | 28 | 100.000% |
| AMB2 | STF2 | 2.371km | G | 40 | 34 | 5 | 97.500% |
| CLFD | CLFE | 3.623km | G | 36 | 36 | - | 100.0% |
| CLFD | OPME | 5.600km | G | 50 | 34 | 16 | 100.000% |
| BRMF | VOUR | 14.218km | G | 38 | 34 | 4 | 100.000% |
| FIED | LONS | 15.984km | G | 64 | 30 | 29 | 92.188% |
| NIVO | STAZ | 17.097km | G | 48 | 38 | 6 | 91.667% |
| MRON | PRNY | 17.571km | G | 54 | 34 | 17 | 94.444% |
| FIED | VAUD | 24.129km | G | 70 | 30 | - | 42.9% |
| SJPL | STGN | 27.356km | G | 40 | 34 | - | 85.0% |
| CBRY | STAZ | 30.124km | G | 48 | 38 | - | 79.2% |
| OPME | SJPL | 31.660km | G | 52 | 38 | - | 73.1% |
| RMZT | SOLR | 32.558km | G | 44 | 36 | - | 81.8% |
| CBRY | CROL | 33.780km | G | 42 | 36 | - | 85.7% |
| BRMF | NIVO | 34.630km | G | 46 | 34 | - | 73.9% |
| RMZT | VISN | 34.746km | G | 36 | 34 | - | 94.4% |
| CROL | VILD | 35.288km | G | 36 | 34 | - | 94.4% |
| DROU | SEUR | 35.428km | G | 44 | 38 | - | 86.4% |
| SEUR | VAUD | 36.191km | G | 38 | 34 | - | 89.5% |
| ANNY | MARG | 37.064km | G | 36 | 34 | - | 94.4% |
| ANAY | UNIX | 37.412km | G | 42 | 36 | - | 85.7% |
| ANNY | CBRY | 38.637km | G | 44 | 34 | - | 77.3% |
| BLG2 | CERN | 38.716km | G | 48 | 34 | - | 70.8% |
| CERN | MARG | 39.704km | G | 44 | 36 | - | 81.8% |
| SOLR | VILD | 41.742km | G | 36 | 32 | - | 88.9% |
| AMB2 | UNIX | 42.944km | G | 46 | 36 | - | 78.3% |
| STGN | VERN | 45.604km | G | 42 | 38 | - | 90.5% |
| ANAY | GLRA | 46.276km | G | 46 | 34 | - | 73.9% |
| ANAY | VOUR | 47.459km | G | 42 | 34 | - | 81.0% |
| AUTN | DROU | 47.925km | G | 44 | 36 | - | 81.8% |
| FIED | MRON | 48.802km | G | 60 | 28 | - | 46.7% |
| PUYV | UNIX | 49.694km | G | 48 | 40 | - | 83.3% |
| STF2 | VERN | 52.434km | G | 42 | 36 | - | 85.7% |
| BLG2 | LONS | 56.342km | G | 34 | 32 | - | 94.1% |
| CLFE | STPS | 60.263km | G | 34 | 34 | - | 100.0% |
| AMB2 | RNNE | 61.026km | G | 42 | 34 | - | 81.0% |
| CROL | MODA | 64.868km | G | 38 | 34 | - | 89.5% |




Fixation des ambiguités Core
| Station 1 | Station 2 | Longueur | Constellation | Ambiguités posées | Ambiguités fixées QIF | Ambiguités fixées L12 | Ratio |
|---|---|---|---|---|---|---|---|
| GRAS | NICA | 25.389km | G | 52 | 30 | - | 57.7% |
| MLVL | TGRI | 46.866km | G | 38 | 34 | - | 89.5% |
| TGRI | TRBL | 54.291km | G | 36 | 34 | - | 94.4% |
| DGLG | LIL2 | 69.994km | G | 50 | 30 | - | 60.0% |
| EGLT | LGES | 78.902km | G | 38 | 32 | - | 84.2% |
| CLFD | EGLT | 91.721km | G | 44 | 32 | - | 72.7% |
| MLVL | ROMY | 94.021km | G | 38 | 34 | - | 89.5% |
| LUMI | SARS | 94.978km | G | 48 | 34 | - | 70.8% |
| ROMY | TRYS | 99.431km | G | 68 | 36 | - | 52.9% |
| CLFD | PUYV | 99.856km | G | 44 | 38 | - | 86.4% |
| CRAL | TLSE | 102.285km | G | 50 | 38 | - | 76.0% |
| AIGL | PUYV | 105.192km | G | 56 | 38 | - | 67.9% |
| MAN2 | TRBL | 106.913km | G | 44 | 34 | - | 77.3% |
| BRMF | PUYV | 112.426km | G | 40 | 36 | - | 90.0% |
| ANGL | STNZ | 113.782km | G | 44 | 36 | - | 81.8% |
| AXPV | GRAS | 131.440km | G | 44 | 32 | - | 72.7% |
| AUTN | BSCN | 133.134km | G | 60 | 38 | - | 63.3% |
| ENTZ | THNV | 140.669km | G | 36 | 32 | - | 88.9% |
| MAN2 | VFCH | 142.465km | G | 42 | 34 | - | 81.0% |
| AUTN | BRMF | 145.348km | G | 42 | 32 | - | 76.2% |
| AUTN | TRYS | 150.482km | G | 72 | 38 | - | 52.8% |
| GENO | NICA | 157.322km | G | 44 | 32 | - | 72.7% |
| AIGL | AXPV | 157.424km | G | 48 | 36 | - | 75.0% |
| LIL2 | ROMY | 166.466km | G | 46 | 30 | - | 65.2% |
| CUBX | LGES | 175.940km | G | 38 | 34 | - | 89.5% |
| ROMY | THNV | 176.050km | G | 54 | 34 | - | 63.0% |
| LUMI | NICA | 178.548km | G | 46 | 26 | - | 56.5% |
| AIGL | TLSE | 180.053km | G | 52 | 34 | - | 65.4% |
| ANGL | CUBX | 182.908km | G | 44 | 32 | - | 72.7% |
| BRST | STNZ | 211.442km | G | 46 | 34 | - | 73.9% |




Estimation des ZTD finaux (coordonnées fixées) - ADDNEQ2
| Statistiques | |
|---|---|
| Paramètres explicites | 9207 |
| Paramètres implicites | 1607 |
| Paramètres ajustés | 10814 |
| Observations | 291187 |
| Degré de liberté | 280373 |
| Chi**2/DOF | 1.86 |
| Stations | 0 |
| Fichier | Début | Fin | Obs. | Par. | Dof |
|---|---|---|---|---|---|
| MET_26069P_1.NQ0 | 2026-03-10 12:00:00 | 2026-03-11 00:00:00 | 30185 | 2208 | 27977 |
| MET_26069P_2.NQ0 | 2026-03-10 12:00:00 | 2026-03-11 00:00:00 | 30706 | 2195 | 28511 |
| MET_26069P_3.NQ0 | 2026-03-10 12:00:00 | 2026-03-11 00:00:00 | 32815 | 2276 | 30539 |
| MET_26069P_4.NQ0 | 2026-03-10 12:00:00 | 2026-03-11 00:00:00 | 32873 | 2300 | 30573 |
| MET_26069P_5.NQ0 | 2026-03-10 12:00:00 | 2026-03-11 00:00:00 | 33813 | 2404 | 31409 |
| MET_26069P_6.NQ0 | 2026-03-10 12:00:00 | 2026-03-11 00:00:00 | 33863 | 2257 | 31606 |
| MET_26069P_7.NQ0 | 2026-03-10 12:00:00 | 2026-03-11 00:00:00 | 32900 | 2346 | 30554 |
| MET_26069P_8.NQ0 | 2026-03-10 12:00:00 | 2026-03-11 00:00:00 | 30772 | 2161 | 28611 |
| MET_26069P_9.NQ0 | 2026-03-10 12:00:00 | 2026-03-11 00:00:00 | 26132 | 1885 | 24247 |