Rapport de calcul
Carte Horaire
Compte rendu de calcul Bernese GPS Software
 Informations sur le processus
Informations sur le processus Sessions intégrées au calcul
Sessions intégrées au calcul Orbites et coordonnées du pôle
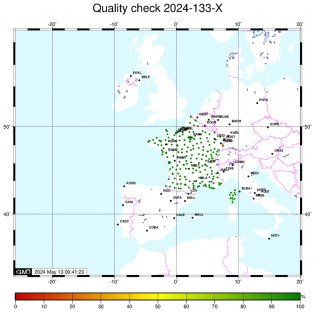
Orbites et coordonnées du pôle Controle qualité
Controle qualité Calcul sur le code
Calcul sur le code Fixation des ambiguités
Fixation des ambiguités Fixation des ambiguités Core
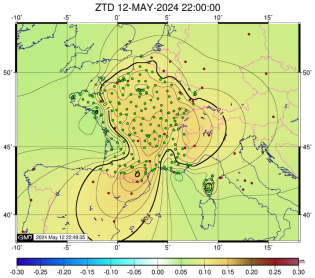
Fixation des ambiguités Core Estimation des ZTD finaux (coordonnées fixées) - ADDNEQ2
Estimation des ZTD finaux (coordonnées fixées) - ADDNEQ2 Estimation des coordonnées et des ZTD finaux - ADDNEQ2
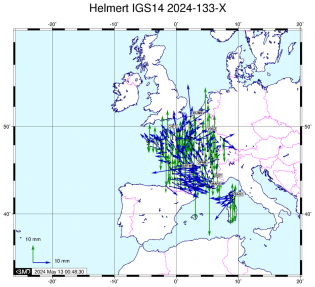
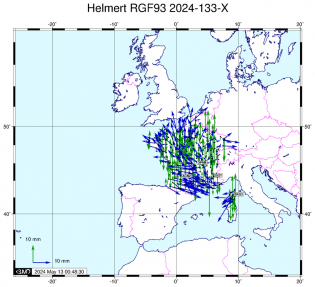
Estimation des coordonnées et des ZTD finaux - ADDNEQ2 Mise en référence IGS14
Mise en référence IGS14



Sessions intégrées au calcul
| Année | DOY | Session |
|---|---|---|
| 2025 | 181 | o |
| 2025 | 181 | p |
| 2025 | 181 | q |
| 2025 | 181 | r |
| 2025 | 181 | s |
| 2025 | 181 | t |




Orbites et coordonnées du pôle
| Orbites | Coordonnées du pôle |
|---|---|
| IGS0OPSULT_20251801200_02D_15M_ORB.SP3.gz | IGS0OPSULT_20251801200_02D_01D_ERP.ERP.gz |




Controle qualité
| Station | Dernier fichier | Ratio des fichiers précédents | Ratio qc_gps | Ratio qc_glo | qc - header GPS | qc - header GLO | IOD slips <10° | IOD slips >10° | Station utilisée dans le calcul |
|---|---|---|---|---|---|---|---|---|---|
| ALBM | - | 0.0% | - | - | - | - | - | - | - |
| AUBU | - | 0.0% | - | - | - | - | - | - | - |
| BRET | - | 0.0% | - | - | - | - | - | - | - |
| CABW | - | 0.0% | - | - | - | - | - | - | - |
| CANT | - | 100.0% | - | - | - | - | - | - | - |
| DOUR | - | 0.0% | - | - | - | - | - | - | - |
| MENS | - | 40.0% | - | - | - | - | - | - | - |
| MZAN | - | 0.0% | - | - | - | - | - | - | - |
| PALI | - | 0.0% | - | - | - | - | - | - | - |
| PANA | - | 40.0% | - | - | - | - | - | - | - |
| PUYA | - | 100.0% | - | - | - | - | - | - | - |
| RIO1 | - | 100.0% | - | - | - | - | - | - | - |
| SEOL | - | 0.0% | - | - | - | - | - | - | - |
| SJDV | - | 0.0% | - | - | - | - | - | - | - |
| STGN | - | 0.0% | - | - | - | - | - | - | - |
| SWAS | - | 0.0% | - | - | - | - | - | - | - |
| THOR | - | 0.0% | - | - | - | - | - | - | - |
| WARE | - | 0.0% | - | - | - | - | - | - | - |
| BLAY | x | 60.0% | 39.2% | - | 3.7002m | - | 0 | 0 | - |
| JANU | x | 80.0% | 68.6% | - | 5.6921m | - | 16 | 7 | x |
| BOUG | x | 80.0% | 79.4% | - | 7.2342m | - | 5 | 1 | x |
| GRAS | x | 100.0% | 83.4% | - | 7.0554m | - | 86 | 18 | x |
| FIED | x | 100.0% | 87.5% | - | 4.9627m | - | 59 | 58 | x |
| RMZT | x | 100.0% | 88.1% | - | 3.1200m | - | 0 | 1 | x |
| STNZ | x | 100.0% | 88.8% | - | 5.4721m | - | 9 | 3 | x |
| SIRT | x | 100.0% | 89.1% | - | 7.5608m | - | 0 | 0 | x |
| FILF | x | 100.0% | 89.6% | - | 3.2998m | - | 7 | 5 | x |
| LOYT | x | 100.0% | 90.3% | - | 3.2041m | - | 0 | 0 | x |
| CDBC | x | 100.0% | 90.5% | - | 7.5591m | - | 31 | 12 | x |
| EXLC | x | 100.0% | 90.5% | - | 10.7592m | - | 44 | 17 | x |
| LAHE | x | 100.0% | 91.8% | - | 5.6689m | - | 298 | 53 | x |
| DEQU | x | 100.0% | 92% | - | 3.7908m | - | 0 | 11 | x |
| VLG2 | x | 100.0% | 92.9% | - | 3.0290m | - | 0 | 0 | x |
| CHTG | x | 100.0% | 93.1% | - | 10.7596m | - | 185 | 12 | x |
| LACA | x | 100.0% | 93.9% | - | 5.4497m | - | 87 | 11 | x |
| PLST | x | 100.0% | 94.4% | - | 4.3225m | - | 0 | 0 | x |
| BUSI | x | 100.0% | 94.6% | - | 6.4711m | - | 0 | 4 | x |
| DHUI | x | 100.0% | 94.9% | - | 3.6861m | - | 0 | 12 | x |
| 1MEL | x | 100.0% | 95.1% | - | 3.4482m | - | 0 | 9 | x |
| COMO | x | 100.0% | 95.1% | - | 7.7347m | - | 5 | 0 | x |
| JOUX | x | 100.0% | 95.1% | - | 6.6493m | - | 28 | 5 | x |
| SIST | x | 100.0% | 95.2% | - | 5.3908m | - | 0 | 2 | x |
| BADH | x | 100.0% | 95.3% | - | 13.7166m | - | 58 | 15 | x |
| JOUS | x | 100.0% | 95.3% | - | 3.2430m | - | 0 | 0 | x |
| BSCN | x | 100.0% | 95.4% | - | 8.2368m | - | 73 | 30 | x |
| DIPP | x | 100.0% | 95.4% | - | 8.0193m | - | 121 | 32 | x |
| VNNS | x | 100.0% | 95.4% | - | 3.6561m | - | 0 | 0 | x |
| HERB | x | 100.0% | 95.7% | - | 4.3882m | - | 0 | 3 | x |
| PARD | x | 100.0% | 95.7% | - | 7.1403m | - | 59 | 3 | x |
| THNV | x | 100.0% | 95.7% | - | 3.2461m | - | 0 | 0 | x |
| PUYV | x | 100.0% | 95.8% | - | 8.8359m | - | 24 | 15 | x |
| BRTM | x | 100.0% | 95.9% | - | 3.3745m | - | 0 | 0 | x |
| MARS | x | 100.0% | 96% | - | 8.3744m | - | 60 | 1 | x |
| MICH | x | 100.0% | 96% | - | 6.4000m | - | 26 | 5 | x |
| FRGN | x | 100.0% | 96.1% | - | 4.5105m | - | 0 | 4 | x |
| DILL | x | 100.0% | 96.3% | - | 7.0507m | - | 49 | 16 | x |
| PLMG | x | 100.0% | 96.3% | - | 5.2339m | - | 0 | 9 | x |
| RUPT | x | 100.0% | 96.3% | - | 4.8382m | - | 0 | 0 | x |
| DOMP | x | 100.0% | 96.4% | - | 4.3395m | - | 0 | 0 | x |
| LMDM | x | 100.0% | 96.4% | - | 3.8534m | - | 0 | 0 | x |
| ATH2 | x | 100.0% | 96.5% | - | 4.3788m | - | 0 | 4 | x |
| BAUG | x | 100.0% | 96.5% | - | 3.2658m | - | 0 | 0 | x |
| LIZ2 | x | 100.0% | 96.5% | - | 6.1749m | - | 0 | 1 | x |
| LMGS | x | 100.0% | 96.5% | - | 3.2777m | - | 0 | 1 | x |
| BLVR | x | 100.0% | 96.6% | - | 7.3588m | - | 62 | 7 | x |
| CLMT | x | 100.0% | 96.6% | - | 5.4021m | - | 0 | 2 | x |
| CONN | x | 100.0% | 96.6% | - | 5.6638m | - | 0 | 0 | x |
| CRTS | x | 100.0% | 96.6% | - | 3.9401m | - | 0 | 0 | x |
| CYLM | x | 100.0% | 96.6% | - | 3.5392m | - | 0 | 6 | x |
| FJC2 | x | 100.0% | 96.6% | - | 7.3986m | - | 11 | 3 | x |
| LESA | x | 100.0% | 96.6% | - | 4.9080m | - | 0 | 0 | x |
| TRBL | x | 100.0% | 96.6% | - | 4.2120m | - | 0 | 23 | x |
| AGDS | x | 100.0% | 96.7% | - | 6.3401m | - | 0 | 0 | x |
| BELL | x | 100.0% | 96.7% | - | 7.2967m | - | 152 | 26 | x |
| BEUG | x | 100.0% | 96.7% | - | 6.8277m | - | 0 | 11 | x |
| BRM2 | x | 100.0% | 96.7% | - | 4.7157m | - | 0 | 1 | x |
| COCR | x | 100.0% | 96.7% | - | 4.8942m | - | 0 | 2 | x |
| GAJN | x | 100.0% | 96.7% | - | 2.4674m | - | 0 | 2 | x |
| MOFA | x | 100.0% | 96.7% | - | 5.7147m | - | 0 | 2 | x |
| ALU2 | x | 100.0% | 96.8% | - | 5.7118m | - | 0 | 0 | x |
| ANDE | x | 100.0% | 96.8% | - | 2.9978m | - | 0 | 0 | x |
| BVOI | x | 100.0% | 96.8% | - | 6.1064m | - | 0 | 1 | x |
| DRUL | x | 100.0% | 96.8% | - | 4.3179m | - | 0 | 3 | x |
| ETOI | x | 100.0% | 96.8% | - | 8.1465m | - | 64 | 23 | x |
| MAGR | x | 100.0% | 96.8% | - | 4.1959m | - | 0 | 0 | x |
| MNBL | x | 100.0% | 96.8% | - | 4.0435m | - | 0 | 0 | x |
| ROMY | x | 100.0% | 96.8% | - | 6.4064m | - | 0 | 0 | x |
| AMNS | x | 100.0% | 96.9% | - | 4.6765m | - | 0 | 0 | x |
| CHE2 | x | 100.0% | 96.9% | - | 5.6014m | - | 0 | 14 | x |
| CNNE | x | 100.0% | 96.9% | - | 7.4853m | - | 28 | 6 | x |
| CRAU | x | 100.0% | 96.9% | - | 4.5418m | - | 0 | 0 | x |
| DBMH | x | 100.0% | 96.9% | - | 6.2605m | - | 0 | 0 | x |
| FEUR | x | 100.0% | 96.9% | - | 3.0901m | - | 0 | 1 | x |
| LRTZ | x | 100.0% | 96.9% | - | 7.1184m | - | 0 | 0 | x |
| MOMO | x | 100.0% | 96.9% | - | 4.9795m | - | 0 | 0 | x |
| NICA | x | 100.0% | 96.9% | - | 6.6342m | - | 0 | 10 | x |
| STAB | x | 100.0% | 96.9% | - | 5.0467m | - | 0 | 0 | x |
| STBR | x | 100.0% | 96.9% | - | 9.0911m | - | 2 | 9 | x |
| TGRI | x | 100.0% | 96.9% | - | 4.2469m | - | 0 | 1 | x |
| TRYS | x | 100.0% | 96.9% | - | 4.3294m | - | 0 | 9 | x |
| VOUZ | x | 100.0% | 96.9% | - | 6.1722m | - | 0 | 4 | x |
| YSCN | x | 100.0% | 96.9% | - | 5.8754m | - | 0 | 0 | x |
| AGDE | x | 100.0% | 97% | - | 6.5246m | - | 77 | 1 | x |
| APT2 | x | 100.0% | 97% | - | 2.8761m | - | 0 | 1 | x |
| BAL2 | x | 100.0% | 97% | - | 5.1213m | - | 0 | 2 | x |
| BOVE | x | 100.0% | 97% | - | 6.5229m | - | 0 | 0 | x |
| BVS2 | x | 100.0% | 97% | - | 6.5645m | - | 0 | 0 | x |
| CHPO | x | 100.0% | 97% | - | 5.4425m | - | 0 | 0 | x |
| DOCX | x | 100.0% | 97% | - | 4.8647m | - | 0 | 0 | x |
| DUNQ | x | 100.0% | 97% | - | 10.3250m | - | 81 | 1 | x |
| EBNS | x | 100.0% | 97% | - | 5.0884m | - | 0 | 0 | x |
| EQH2 | x | 100.0% | 97% | - | 6.8564m | - | 0 | 0 | x |
| GAPC | x | 100.0% | 97% | - | 3.1448m | - | 0 | 0 | x |
| GUIP | x | 100.0% | 97% | - | 8.9972m | - | 74 | 44 | x |
| LCST | x | 100.0% | 97% | - | 4.0979m | - | 0 | 7 | x |
| MATC | x | 100.0% | 97% | - | 3.7060m | - | 0 | 3 | x |
| MIRE | x | 100.0% | 97% | - | 4.2912m | - | 0 | 0 | x |
| NOGT | x | 100.0% | 97% | - | 4.6337m | - | 0 | 0 | x |
| PAMO | x | 100.0% | 97% | - | 3.6015m | - | 0 | 0 | x |
| PRR2 | x | 100.0% | 97% | - | 6.5465m | - | 0 | 0 | x |
| SAMR | x | 100.0% | 97% | - | 5.9994m | - | 0 | 0 | x |
| SNNT | x | 100.0% | 97% | - | 3.7879m | - | 0 | 4 | x |
| STGS | x | 100.0% | 97% | - | 2.5716m | - | 0 | 7 | x |
| VNTE | x | 100.0% | 97% | - | 5.8831m | - | 0 | 0 | x |
| ARNA | x | 100.0% | 97.1% | - | 3.5837m | - | 0 | 0 | x |
| BAPI | x | 100.0% | 97.1% | - | 4.3704m | - | 0 | 1 | x |
| BARD | x | 100.0% | 97.1% | - | 3.7309m | - | 0 | 0 | x |
| BRST | x | 100.0% | 97.1% | - | 8.2735m | - | 62 | 5 | x |
| CANE | x | 100.0% | 97.1% | - | 8.6198m | - | 2 | 0 | x |
| CCSL | x | 100.0% | 97.1% | - | 4.3698m | - | 0 | 9 | x |
| CH2T | x | 100.0% | 97.1% | - | 5.9316m | - | 0 | 0 | x |
| CULA | x | 100.0% | 97.1% | - | 4.4484m | - | 0 | 0 | x |
| DGLG | x | 100.0% | 97.1% | - | 8.7945m | - | 145 | 8 | x |
| GRAC | x | 100.0% | 97.1% | - | 8.2365m | - | 110 | 0 | x |
| LAVL | x | 100.0% | 97.1% | - | 5.4168m | - | 0 | 4 | x |
| LCVA | x | 100.0% | 97.1% | - | 3.1511m | - | 0 | 1 | x |
| LEZI | x | 100.0% | 97.1% | - | 3.2464m | - | 0 | 1 | x |
| MEDI | x | 100.0% | 97.1% | - | 9.5371m | - | 171 | 13 | x |
| MONS | x | 100.0% | 97.1% | - | 4.1453m | - | 0 | 0 | x |
| MTLC | x | 100.0% | 97.1% | - | 2.8703m | - | 0 | 0 | x |
| NOYL | x | 100.0% | 97.1% | - | 4.8324m | - | 0 | 0 | x |
| PLGN | x | 100.0% | 97.1% | - | 5.4927m | - | 0 | 0 | x |
| PRAD | x | 100.0% | 97.1% | - | 2.6587m | - | 0 | 0 | x |
| PTRC | x | 100.0% | 97.1% | - | 3.8113m | - | 0 | 0 | x |
| SAFQ | x | 100.0% | 97.1% | - | 2.6954m | - | 0 | 0 | x |
| STF2 | x | 100.0% | 97.1% | - | 3.6079m | - | 0 | 0 | x |
| TLTG | x | 100.0% | 97.1% | - | 10.8343m | - | 29 | 0 | x |
| UZER | x | 100.0% | 97.1% | - | 4.7084m | - | 0 | 0 | x |
| VEN2 | x | 100.0% | 97.1% | - | 4.2616m | - | 0 | 0 | x |
| VLPL | x | 100.0% | 97.1% | - | 4.6861m | - | 0 | 0 | x |
| AAER | x | 100.0% | 97.2% | - | 7.1777m | - | 25 | 0 | x |
| AICI | x | 100.0% | 97.2% | - | 2.2248m | - | 0 | 1 | x |
| ALGY | x | 100.0% | 97.2% | - | 4.4903m | - | 0 | 1 | x |
| AMIY | x | 100.0% | 97.2% | - | 4.8264m | - | 0 | 4 | x |
| AUBT | x | 100.0% | 97.2% | - | 3.1888m | - | 0 | 0 | x |
| AUT2 | x | 100.0% | 97.2% | - | 3.8077m | - | 0 | 1 | x |
| BRE2 | x | 100.0% | 97.2% | - | 2.2472m | - | 0 | 0 | x |
| BRG9 | x | 100.0% | 97.2% | - | 4.5067m | - | 0 | 0 | x |
| BXME | x | 100.0% | 97.2% | - | 8.7315m | - | 47 | 8 | x |
| C2CN | x | 100.0% | 97.2% | - | 5.2850m | - | 0 | 0 | x |
| CABN | x | 100.0% | 97.2% | - | 3.2580m | - | 0 | 1 | x |
| CAMO | x | 100.0% | 97.2% | - | 10.8721m | - | 14 | 0 | x |
| CHBL | x | 100.0% | 97.2% | - | 3.2025m | - | 0 | 0 | x |
| CHIO | x | 100.0% | 97.2% | - | 6365348.9032m | - | 44 | 0 | x |
| CHOL | x | 100.0% | 97.2% | - | 4.1195m | - | 0 | 0 | x |
| CLFD | x | 100.0% | 97.2% | - | 5.2498m | - | 0 | 0 | x |
| CUBX | x | 100.0% | 97.2% | - | 7.7750m | - | 37 | 3 | x |
| ENTZ | x | 100.0% | 97.2% | - | 9.4621m | - | 34 | 0 | x |
| EOST | x | 100.0% | 97.2% | - | 8.6825m | - | 49 | 1 | x |
| ESCO | x | 100.0% | 97.2% | - | 7.0842m | - | 27 | 0 | x |
| GDIJ | x | 100.0% | 97.2% | - | 3.3871m | - | 1 | 0 | x |
| GIE1 | x | 100.0% | 97.2% | - | 4.2338m | - | 0 | 0 | x |
| KEGA | x | 100.0% | 97.2% | - | 3.6387m | - | 0 | 1 | x |
| LAIG | x | 100.0% | 97.2% | - | 5.9411m | - | 0 | 0 | x |
| LDB2 | x | 100.0% | 97.2% | - | 7.3334m | - | 71 | 1 | x |
| LGES | x | 100.0% | 97.2% | - | 4.1450m | - | 0 | 3 | x |
| LHVE | x | 100.0% | 97.2% | - | 6.4281m | - | 0 | 1 | x |
| LIL2 | x | 100.0% | 97.2% | - | 7.2371m | - | 59 | 0 | x |
| LLGV | x | 100.0% | 97.2% | - | 3.8898m | - | 0 | 0 | x |
| MAUP | x | 100.0% | 97.2% | - | 3.8263m | - | 0 | 0 | x |
| MEND | x | 100.0% | 97.2% | - | 5.0259m | - | 0 | 0 | x |
| MPRV | x | 100.0% | 97.2% | - | 4.5016m | - | 0 | 0 | x |
| MTIL | x | 100.0% | 97.2% | - | 3.0447m | - | 0 | 0 | x |
| NMTR | x | 100.0% | 97.2% | - | 4.0078m | - | 0 | 0 | x |
| NVPT | x | 100.0% | 97.2% | - | 4.2377m | - | 0 | 2 | x |
| OMBA | x | 100.0% | 97.2% | - | 4.3895m | - | 0 | 0 | x |
| OPME | x | 100.0% | 97.2% | - | 4.2964m | - | 0 | 2 | x |
| PEY2 | x | 100.0% | 97.2% | - | 3.0957m | - | 0 | 2 | x |
| PLDN | x | 100.0% | 97.2% | - | 5.0091m | - | 0 | 3 | x |
| PNTV | x | 100.0% | 97.2% | - | 4.3793m | - | 0 | 0 | x |
| QNSN | x | 100.0% | 97.2% | - | 2.7065m | - | 0 | 1 | x |
| QUYC | x | 100.0% | 97.2% | - | 2.8871m | - | 0 | 0 | x |
| RGNC | x | 100.0% | 97.2% | - | 4.4576m | - | 0 | 2 | x |
| RYO2 | x | 100.0% | 97.2% | - | 4.5552m | - | 0 | 0 | x |
| SAUN | x | 100.0% | 97.2% | - | 3.4333m | - | 0 | 0 | x |
| SDDP | x | 100.0% | 97.2% | - | 4.9335m | - | 0 | 0 | x |
| SHOE | x | 100.0% | 97.2% | - | 6365118.8774m | - | 47 | 0 | x |
| SMCR | x | 100.0% | 97.2% | - | 4.2984m | - | 0 | 0 | x |
| SPIO | x | 100.0% | 97.2% | - | 3.4610m | - | 0 | 0 | x |
| STAZ | x | 100.0% | 97.2% | - | 3.5599m | - | 0 | 0 | x |
| STEL | x | 100.0% | 97.2% | - | 5.7752m | - | 0 | 0 | x |
| STMA | x | 100.0% | 97.2% | - | 4.6955m | - | 0 | 0 | x |
| VAUD | x | 100.0% | 97.2% | - | 4.8166m | - | 0 | 0 | x |
| VERN | x | 100.0% | 97.2% | - | 3.2133m | - | 0 | 0 | x |
| VIN1 | x | 100.0% | 97.2% | - | 3.7848m | - | 0 | 0 | x |
| VLIS | x | 100.0% | 97.2% | - | 9.3306m | - | 50 | 0 | x |
| VRCT | x | 100.0% | 97.2% | - | 3.5609m | - | 0 | 1 | x |
| AGN2 | x | 100.0% | 97.3% | - | 3.0190m | - | 0 | 0 | x |
| ARDN | x | 100.0% | 97.3% | - | 3.9185m | - | 0 | 0 | x |
| ASNE | x | 100.0% | 97.3% | - | 3.4154m | - | 0 | 0 | x |
| AUNI | x | 100.0% | 97.3% | - | 3.7614m | - | 0 | 0 | x |
| CAPT | x | 100.0% | 97.3% | - | 3.7542m | - | 0 | 0 | x |
| CORZ | x | 100.0% | 97.3% | - | 2.4231m | - | 0 | 0 | x |
| CREU | x | 100.0% | 97.3% | - | 8.3883m | - | 269 | 0 | x |
| CSTO | x | 100.0% | 97.3% | - | 2.2247m | - | 0 | 0 | x |
| EGLT | x | 100.0% | 97.3% | - | 6.8255m | - | 49 | 9 | x |
| LHGE | x | 100.0% | 97.3% | - | 5.7634m | - | 0 | 0 | x |
| MAZE | x | 100.0% | 97.3% | - | 3.7322m | - | 0 | 0 | x |
| NMCU | x | 100.0% | 97.3% | - | 5.4524m | - | 5 | 0 | x |
| SCIL | x | 100.0% | 97.3% | - | 6365757.9487m | - | 23 | 0 | x |
| SCOA | x | 100.0% | 97.3% | - | 10.7304m | - | 118 | 25 | x |
| SEM2 | x | 100.0% | 97.3% | - | 3.1515m | - | 0 | 1 | x |
| SMDP | x | 100.0% | 97.3% | - | 3.3370m | - | 0 | 0 | x |
| STMR | x | 100.0% | 97.3% | - | 5.0958m | - | 107 | 1 | x |
| STS2 | x | 100.0% | 97.3% | - | 5.3967m | - | 0 | 0 | x |
| TANC | x | 100.0% | 97.3% | - | 7.9372m | - | 118 | 7 | x |
| VSOU | x | 100.0% | 97.3% | - | 3.1370m | - | 0 | 0 | x |
| AUTN | x | 100.0% | 97.4% | - | 7.5261m | - | 33 | 4 | x |
| AXPV | x | 100.0% | 97.4% | - | 10.1338m | - | 71 | 0 | x |
| BANN | x | 100.0% | 97.4% | - | 8.2366m | - | 54 | 0 | x |
| BRMF | x | 100.0% | 97.4% | - | 8.3722m | - | 56 | 0 | x |
| CERN | x | 100.0% | 97.4% | - | 7.9784m | - | 16 | 4 | x |
| CNTL | x | 100.0% | 97.4% | - | 9.8796m | - | 85 | 1 | x |
| CRAL | x | 100.0% | 97.4% | - | 7.0534m | - | 12 | 0 | x |
| EBRE | x | 100.0% | 97.4% | - | 7.0247m | - | 82 | 1 | x |
| EXSG | x | 100.0% | 97.4% | - | 7.5699m | - | 20 | 0 | x |
| FAJP | x | 100.0% | 97.4% | - | 6.6958m | - | 9 | 0 | x |
| FRAN | x | 100.0% | 97.4% | - | 9.1766m | - | 235 | 2 | x |
| GARD | x | 100.0% | 97.4% | - | 3.5660m | - | 0 | 0 | x |
| GRIG | x | 100.0% | 97.4% | - | 8.0032m | - | 8 | 0 | x |
| HOLA | x | 100.0% | 97.4% | - | 7.9077m | - | 60 | 1 | x |
| HONF | x | 100.0% | 97.4% | - | 8.5077m | - | 102 | 1 | x |
| HOUL | x | 100.0% | 97.4% | - | 8.4297m | - | 157 | 0 | x |
| IGNP | x | 100.0% | 97.4% | - | 10.6828m | - | 8 | 0 | x |
| ILDX | x | 100.0% | 97.4% | - | 8.9988m | - | 69 | 0 | x |
| LBTM | x | 100.0% | 97.4% | - | 9.3722m | - | 28 | 1 | x |
| MAN2 | x | 100.0% | 97.4% | - | 8.1111m | - | 55 | 1 | x |
| MLVL | x | 100.0% | 97.4% | - | 7.0049m | - | 37 | 0 | x |
| NGER | x | 100.0% | 97.4% | - | 2.6021m | - | 0 | 0 | x |
| PAYE | x | 100.0% | 97.4% | - | 8.8185m | - | 78 | 5 | x |
| SBGS | x | 100.0% | 97.4% | - | 7.5207m | - | 2 | 0 | x |
| SET1 | x | 100.0% | 97.4% | - | 7.5271m | - | 87 | 0 | x |
| STJF | x | 100.0% | 97.4% | - | 9.4066m | - | 71 | 0 | x |
| TLMF | x | 100.0% | 97.4% | - | 9.5425m | - | 120 | 0 | x |
| TLSE | x | 100.0% | 97.4% | - | 6.5067m | - | 48 | 0 | x |
| TLSG | x | 100.0% | 97.4% | - | 8.1212m | - | 19 | 0 | x |
| VFCH | x | 100.0% | 97.4% | - | 9.7979m | - | 45 | 0 | x |




Calcul des coordonnées sur le code
Les stations sont éliminées du calcul si l'écart entre les coordonnées issues du calcul sur le code et les coordonnées initiales est supérieur à dE=10.0 m, dN=10.0 m ou dU=20.0 m| Station | dE (m) | dN (m) | dU (m) |
|---|---|---|---|
| 1MEL | 0.14m | -0.15m | -0.06m |
| AAER | -0.22m | -0.18m | 0.70m |
| AGDE | 0.10m | 0.12m | 0.54m |
| AGDS | 0.06m | 0.23m | -0.14m |
| AGN2 | 0.09m | 0.03m | -0.14m |
| AICI | 0.12m | 0.18m | -0.16m |
| ALGY | -0.12m | 0.12m | 0.27m |
| ALU2 | 0.11m | 0.10m | 0.52m |
| AMIY | -0.01m | 0.03m | 0.12m |
| AMNS | -0.04m | 0.06m | -0.07m |
| ANDE | 0.00m | -0.03m | 0.44m |
| APT2 | 0.15m | 0.14m | -0.55m |
| ARDN | 0.03m | 0.10m | -0.27m |
| ARNA | -0.02m | 0.08m | -0.08m |
| ASNE | 0.10m | 0.02m | 0.02m |
| ATH2 | -0.07m | 0.07m | 0.71m |
| AUBT | -0.02m | 0.24m | -0.19m |
| AUNI | 0.10m | 0.15m | 0.22m |
| AUT2 | -0.12m | 0.08m | 0.22m |
| AUTN | -0.27m | -0.04m | 0.47m |
| AXPV | -0.20m | -0.08m | 0.49m |
| BADH | -0.18m | 0.01m | 0.72m |
| BAL2 | -0.07m | -0.03m | 0.80m |
| BANN | -0.08m | -0.01m | 1.11m |
| BAPI | 0.04m | -0.06m | -0.12m |
| BARD | -0.08m | 0.26m | -0.20m |
| BAUG | 0.05m | 0.18m | 0.05m |
| BELL | -0.02m | 0.02m | 0.78m |
| BEUG | -0.06m | 0.21m | 0.27m |
| BLVR | -0.12m | 0.13m | 1.39m |
| BOUG | 0.48m | 3.78m | -28.57m |
| BOVE | -0.06m | 0.19m | 0.22m |
| BRE2 | 0.02m | 0.03m | -0.04m |
| BRG9 | -0.09m | 0.13m | -0.13m |
| BRM2 | -0.16m | 0.03m | 0.60m |
| BRMF | -0.26m | -0.01m | 1.03m |
| BRST | -0.08m | -0.00m | 0.22m |
| BRTM | 0.11m | 0.09m | -0.58m |
| BSCN | -0.08m | 0.13m | 0.49m |
| BUSI | -0.05m | 0.17m | 0.24m |
| BVOI | -0.04m | -0.03m | 0.44m |
| BVS2 | 0.13m | 0.17m | 0.42m |
| BXME | -0.19m | 0.14m | 0.65m |
| C2CN | 0.05m | 0.24m | -0.17m |
| CABN | 0.15m | 0.10m | 0.09m |
| CAMO | -0.09m | -0.10m | 1.03m |
| CANE | 0.03m | 0.20m | 0.74m |
| CAPT | -0.09m | 0.06m | -0.06m |
| CCSL | -0.07m | -0.01m | 0.07m |
| CDBC | 0.09m | 0.19m | 0.41m |
| CERN | -0.03m | -0.23m | 0.80m |
| CH2T | 0.03m | 0.16m | 0.07m |
| CHBL | 0.15m | 0.10m | -0.88m |
| CHE2 | 0.01m | 0.06m | 0.12m |
| CHIO | 0.01m | 0.01m | 1.10m |
| CHOL | 0.04m | 0.19m | 0.19m |
| CHPO | -0.10m | 0.06m | 0.59m |
| CHTG | -0.11m | 0.32m | 1.32m |
| CLFD | -0.08m | 0.24m | 0.28m |
| CLMT | 0.01m | 0.33m | -0.12m |
| CNNE | 0.09m | 0.06m | 0.74m |
| CNTL | -0.13m | -0.02m | 0.57m |
| COCR | -0.30m | 0.55m | 0.51m |
| COMO | 0.27m | 0.17m | 0.27m |
| CONN | 0.00m | 0.67m | 0.26m |
| CORZ | -0.06m | -0.08m | -0.31m |
| CRAL | 0.08m | 0.22m | 0.33m |
| CRAU | 0.07m | 0.12m | -0.01m |
| CREU | -0.02m | -0.27m | 0.10m |
| CRTS | 0.12m | 0.03m | 0.42m |
| CSTO | 0.06m | 0.05m | -0.11m |
| CUBX | -0.07m | 0.14m | 0.64m |
| CULA | -0.11m | 0.65m | 0.54m |
| CYLM | -0.08m | 0.00m | 0.05m |
| DBMH | -0.03m | 0.15m | 0.42m |
| DEQU | -0.03m | 0.11m | 0.53m |
| DGLG | -0.07m | 0.05m | 0.50m |
| DHUI | -0.12m | -0.02m | 0.22m |
| DILL | -0.18m | -0.07m | 1.08m |
| DIPP | 0.07m | -0.01m | 0.55m |
| DOCX | -0.09m | -0.03m | 0.48m |
| DOMP | 0.12m | -0.04m | 0.45m |
| DRUL | 0.08m | 0.01m | 0.47m |
| DUNQ | -0.07m | 0.23m | 1.10m |
| EBNS | -0.06m | 0.02m | 0.55m |
| EBRE | -0.06m | -0.01m | -0.51m |
| EGLT | -0.06m | -0.01m | 0.26m |
| ENTZ | -0.16m | -0.01m | 0.88m |
| EOST | -0.17m | 0.28m | 1.38m |
| EQH2 | -0.06m | 0.23m | 0.28m |
| ESCO | -0.07m | 0.05m | 0.44m |
| ETOI | -0.05m | -0.11m | 0.92m |
| EXLC | 2.87m | 3.94m | -28.73m |
| EXSG | -0.17m | -0.51m | 0.39m |
| FAJP | -0.15m | 0.11m | 0.13m |
| FEUR | 0.13m | 0.14m | 0.45m |
| FIED | -0.00m | 0.34m | 1.12m |
| FILF | 0.17m | 0.38m | 0.48m |
| FJC2 | -0.14m | 0.26m | 0.12m |
| FRAN | -0.31m | -0.38m | 1.27m |
| FRGN | -0.18m | 0.64m | 1.04m |
| GAJN | 0.10m | -0.10m | 0.05m |
| GAPC | 0.08m | 0.02m | 0.61m |
| GARD | -0.02m | 0.20m | 0.24m |
| GDIJ | 0.09m | 0.11m | 0.16m |
| GIE1 | -0.01m | 0.02m | 0.23m |
| GRAC | -0.20m | -0.08m | 0.91m |
| GRAS | -0.07m | -0.11m | 1.67m |
| GRIG | -0.04m | -0.14m | 0.23m |
| GUIP | -0.17m | 0.19m | 0.20m |
| HERB | -0.02m | 0.20m | 0.16m |
| HOLA | -0.30m | -0.11m | 1.09m |
| HONF | -0.06m | -0.09m | 0.70m |
| HOUL | -0.01m | 0.03m | 0.61m |
| IGNP | -0.07m | -0.09m | 0.37m |
| ILDX | -0.16m | 0.12m | 0.07m |
| JANU | -0.32m | 0.32m | 0.73m |
| JOUS | 0.05m | 0.12m | -0.20m |
| JOUX | -0.15m | -0.02m | 0.75m |
| KEGA | 0.05m | 0.15m | 0.16m |
| LACA | -0.11m | 0.32m | 1.13m |
| LAHE | 0.01m | 0.02m | 0.43m |
| LAIG | -0.08m | -0.01m | 0.25m |
| LAVL | 0.11m | -0.05m | 0.36m |
| LBTM | -0.11m | 0.13m | 0.17m |
| LCST | -0.01m | -0.04m | 0.34m |
| LCVA | 0.08m | 0.07m | -0.08m |
| LDB2 | 0.03m | -0.06m | 0.49m |
| LESA | 0.03m | 0.16m | -0.27m |
| LEZI | -0.02m | 0.07m | 0.08m |
| LGES | 0.13m | 0.14m | 0.18m |
| LHGE | -0.11m | 0.12m | 0.21m |
| LHVE | 0.04m | 0.36m | 0.42m |
| LIL2 | -0.20m | 0.01m | 0.53m |
| LIZ2 | 0.64m | 0.06m | 0.16m |
| LLGV | -0.13m | -0.01m | 0.42m |
| LMDM | 0.04m | 0.70m | 0.84m |
| LMGS | 0.01m | 0.06m | -0.25m |
| LOYT | -0.10m | 0.06m | -0.21m |
| LRTZ | 0.02m | 0.03m | 0.74m |
| MAGR | -0.25m | 0.49m | 0.64m |
| MAN2 | -0.10m | 0.05m | 0.52m |
| MARS | 0.16m | -0.02m | -0.02m |
| MATC | 0.25m | 0.74m | 0.61m |
| MAUP | 0.05m | 0.05m | 0.49m |
| MAZE | 0.04m | 0.03m | -0.10m |
| MEDI | -0.10m | -0.01m | 0.72m |
| MEND | 0.18m | 0.08m | 0.19m |
| MICH | -0.18m | -0.06m | 1.49m |
| MIRE | 0.00m | -0.04m | 0.47m |
| MLVL | -0.16m | -0.00m | 0.43m |
| MNBL | 0.07m | 0.18m | 0.19m |
| MOFA | -0.03m | 0.15m | 0.29m |
| MOMO | -0.03m | 0.22m | 0.10m |
| MONS | -0.44m | 0.34m | 0.73m |
| MPRV | 0.13m | 0.32m | 0.23m |
| MTIL | 0.04m | -0.12m | -0.56m |
| MTLC | 0.13m | -0.02m | -0.05m |
| NGER | 0.05m | 0.12m | -0.09m |
| NICA | -0.01m | 0.22m | 0.49m |
| NMCU | 0.52m | -0.76m | -0.66m |
| NMTR | -0.00m | 0.24m | -0.30m |
| NOGT | -0.06m | -0.04m | 0.17m |
| NOYL | 0.13m | 0.23m | 0.16m |
| NVPT | -0.12m | -0.06m | 0.34m |
| OMBA | 0.08m | 0.02m | 0.15m |
| OPME | 0.04m | 0.26m | 0.23m |
| PAMO | -0.45m | 0.47m | 0.81m |
| PARD | -0.02m | 0.30m | 0.63m |
| PAYE | -0.27m | -0.06m | 1.21m |
| PEY2 | 0.12m | 0.03m | -0.37m |
| PLDN | -0.03m | 0.24m | -0.51m |
| PLGN | -0.15m | 0.23m | -0.11m |
| PLMG | -0.02m | 0.19m | -0.14m |
| PLST | 0.07m | -0.00m | 0.16m |
| PNTV | -0.07m | 0.14m | -0.02m |
| PRAD | 0.09m | 0.13m | -0.23m |
| PRR2 | 0.06m | 0.35m | 0.03m |
| PTRC | -0.09m | -0.06m | -0.16m |
| PUYV | -0.21m | -0.16m | 0.80m |
| QNSN | -0.22m | 0.12m | 0.55m |
| QUYC | -0.07m | 0.05m | -0.46m |
| RGNC | 0.20m | 0.30m | -0.05m |
| RMZT | -0.04m | 0.05m | 0.27m |
| ROMY | -0.07m | 0.10m | 0.30m |
| RUPT | -0.11m | -0.08m | 0.26m |
| RYO2 | -0.19m | 0.13m | -0.04m |
| SAFQ | 0.12m | -0.00m | 0.15m |
| SAMR | -0.05m | 0.20m | -0.31m |
| SAUN | 0.21m | 0.31m | 0.15m |
| SBGS | -0.01m | -0.12m | 0.17m |
| SCIL | -0.12m | 0.02m | 1.03m |
| SCOA | -0.15m | 0.04m | 0.16m |
| SDDP | -0.00m | 0.22m | -0.07m |
| SEM2 | 0.09m | 0.01m | -0.21m |
| SET1 | -0.23m | -0.10m | 1.02m |
| SHOE | -0.19m | 0.14m | 0.69m |
| SIRT | -0.03m | 0.31m | 0.18m |
| SIST | 0.08m | 0.20m | 0.02m |
| SMCR | 0.01m | 0.09m | 0.19m |
| SMDP | 0.09m | 0.23m | 0.15m |
| SNNT | 0.10m | 0.19m | 0.35m |
| SPIO | -0.03m | 0.05m | -0.37m |
| STAB | -0.05m | 0.18m | 0.27m |
| STAZ | -0.03m | 0.00m | 0.43m |
| STBR | -0.09m | 0.18m | 0.29m |
| STEL | -0.02m | 0.22m | 0.13m |
| STF2 | 0.06m | 0.15m | 0.13m |
| STGS | 0.09m | -0.06m | -0.66m |
| STJF | -0.10m | 0.11m | 0.63m |
| STMA | -0.09m | 0.18m | -0.10m |
| STMR | 0.08m | 0.21m | 0.33m |
| STNZ | 0.64m | -0.67m | -0.23m |
| STS2 | 0.02m | 0.26m | -0.40m |
| TANC | -0.05m | 0.15m | 0.36m |
| TGRI | -0.40m | 0.45m | 0.70m |
| THNV | -0.08m | -0.10m | 0.29m |
| TLMF | -0.09m | 0.00m | -0.16m |
| TLSE | -0.10m | -0.12m | 0.72m |
| TLSG | 0.02m | -0.13m | 0.41m |
| TLTG | -0.09m | 0.00m | 0.32m |
| TRBL | -0.03m | 0.08m | 0.16m |
| TRYS | -0.04m | 0.06m | 0.31m |
| UZER | 0.15m | 0.12m | -0.20m |
| VAUD | -0.08m | 0.09m | 0.49m |
| VEN2 | -0.00m | 0.14m | 0.46m |
| VERN | -0.01m | 0.13m | 0.08m |
| VFCH | -0.13m | 0.10m | 0.00m |
| VIN1 | -0.37m | 0.53m | 0.95m |
| VLG2 | 0.09m | 0.03m | -0.01m |
| VLIS | -0.29m | 0.03m | 0.86m |
| VLPL | 0.27m | 0.02m | -0.28m |
| VNNS | 0.04m | 0.08m | -0.17m |
| VNTE | -0.05m | 0.06m | 0.42m |
| VOUZ | -0.04m | 0.17m | 0.49m |
| VRCT | -0.09m | 0.14m | 0.48m |
| VSOU | -0.05m | 0.04m | 0.49m |
| YSCN | -0.84m | 1.51m | -12.40m |




Fixation des ambiguités
| Station 1 | Station 2 | Longueur | Constellation | Ambiguités posées | Ambiguités fixées QIF | Ambiguités fixées L12 | Ratio |
|---|---|---|---|---|---|---|---|
| ENTZ | EOST | 9.663km | G | 44 | 30 | 9 | 88.636% |
| BRM2 | EOST | 15.180km | G | 58 | 30 | 24 | 93.103% |
| DBMH | LRTZ | 15.960km | G | 50 | 32 | 16 | 96.000% |
| BLVR | MNBL | 24.159km | G | 68 | 20 | - | 29.4% |
| CHPO | VOUZ | 34.709km | G | 64 | 40 | - | 62.5% |
| BLVR | LLGV | 35.928km | G | 64 | 24 | - | 37.5% |
| ALGY | BSCN | 37.489km | G | 66 | 26 | - | 39.4% |
| DILL | THNV | 38.457km | G | 58 | 32 | - | 55.2% |
| AAER | CHPO | 38.616km | G | 46 | 30 | - | 65.2% |
| DBMH | MIRE | 38.768km | G | 60 | 32 | - | 53.3% |
| LLGV | PAYE | 42.066km | G | 56 | 32 | - | 57.1% |
| ANDE | RUPT | 42.514km | G | 66 | 24 | - | 36.4% |
| ATH2 | CHPO | 42.962km | G | 56 | 30 | - | 53.6% |
| VSOU | BSCN | 43.298km | G | 54 | 28 | - | 51.9% |
| LLGV | BSCN | 45.163km | G | 54 | 28 | - | 51.9% |
| ATH2 | ROMY | 45.708km | G | 50 | 28 | - | 56.0% |
| NOGT | RUPT | 46.410km | G | 88 | 26 | - | 29.5% |
| BRM2 | DRUL | 47.902km | G | 60 | 40 | - | 66.7% |
| CONN | ROMY | 50.526km | G | 52 | 30 | - | 57.7% |
| ANDE | BVS2 | 53.413km | G | 54 | 28 | - | 51.9% |
| BVS2 | VOUZ | 54.854km | G | 62 | 34 | - | 54.8% |
| DILL | DRUL | 59.982km | G | 66 | 32 | - | 48.5% |
| BVS2 | THNV | 61.377km | G | 54 | 32 | - | 59.3% |
| MIRE | NOGT | 64.363km | G | 60 | 28 | - | 46.7% |
| ALGY | NOGT | 66.717km | G | 66 | 24 | - | 36.4% |
| BAL2 | ENTZ | 69.165km | G | 52 | 30 | - | 57.7% |
| BADH | DILL | 167.281km | G | 52 | 30 | - | 57.7% |
| BADH | LDB2 | 443.418km | G | 48 | 36 | - | 75.0% |
| EOST | ETOI | 1.053km | G | 48 | 32 | 16 | 100.000% |
| GRAC | GRAS | 0.032km | G | 40 | 36 | 4 | 100.000% |
| CRAU | TLTG | 13.271km | G | 38 | 34 | 2 | 94.737% |
| CANE | GRAC | 23.531km | G | 38 | 30 | - | 78.9% |
| CANE | NICA | 23.758km | G | 52 | 32 | - | 61.5% |
| LESA | STMR | 24.574km | G | 56 | 34 | - | 60.7% |
| APT2 | MICH | 26.974km | G | 56 | 26 | - | 46.4% |
| GAPC | SIST | 31.932km | G | 100 | 36 | - | 36.0% |
| MICH | QNSN | 33.591km | G | 62 | 28 | - | 45.2% |
| YSCN | SET1 | 36.243km | G | 64 | 30 | - | 46.9% |
| GAJN | YSCN | 36.925km | G | 56 | 30 | - | 53.6% |
| MICH | SIST | 37.330km | G | 98 | 36 | - | 36.7% |
| APT2 | AXPV | 43.016km | G | 40 | 28 | - | 70.0% |
| CRTS | RMZT | 43.547km | G | 46 | 32 | - | 69.6% |
| EBNS | STAZ | 46.487km | G | 64 | 30 | - | 46.9% |
| RMZT | SIST | 48.822km | G | 84 | 34 | - | 40.5% |
| BANN | MEND | 52.098km | G | 48 | 28 | - | 58.3% |
| BANN | GAJN | 52.118km | G | 48 | 28 | - | 58.3% |
| AXPV | LESA | 53.042km | G | 48 | 30 | - | 62.5% |
| GAJN | STMR | 53.114km | G | 48 | 30 | - | 62.5% |
| AXPV | TLTG | 62.764km | G | 36 | 32 | - | 88.9% |
| MEND | PUYV | 65.940km | G | 50 | 28 | - | 56.0% |
| GRAS | QNSN | 71.999km | G | 48 | 28 | - | 58.3% |
| CRTS | EBNS | 73.993km | G | 70 | 24 | - | 34.3% |
| DEQU | STAZ | 93.344km | G | 74 | 32 | - | 43.2% |
| COMO | DEQU | 191.721km | G | 62 | 24 | - | 38.7% |
| COMO | MEDI | 246.023km | G | 48 | 32 | - | 66.7% |
| CANE | CNNE | 4.342km | G | 46 | 32 | 1 | 71.739% |
| AXPV | MARS | 23.663km | G | 38 | 30 | - | 78.9% |
| GAPC | JANU | 69.771km | G | 52 | 20 | - | 38.5% |
| AUT2 | AUTN | 2.240km | G | 64 | 30 | 31 | 95.312% |
| CLFD | CLMT | 3.105km | G | 70 | 28 | 36 | 91.429% |
| GDIJ | VRCT | 5.204km | G | 66 | 28 | 32 | 90.909% |
| JOUX | SMCR | 5.332km | G | 52 | 34 | 13 | 90.385% |
| CLFD | OPME | 5.600km | G | 54 | 30 | 19 | 90.741% |
| FIED | VAUD | 24.129km | G | 56 | 20 | - | 35.7% |
| FIED | SMCR | 24.305km | G | 62 | 32 | - | 51.6% |
| ALU2 | STEL | 33.234km | G | 46 | 30 | - | 65.2% |
| CERN | JOUX | 36.430km | G | 62 | 26 | - | 41.9% |
| DOCX | JOUX | 38.125km | G | 56 | 28 | - | 50.0% |
| CERN | MTLC | 38.466km | G | 56 | 30 | - | 53.6% |
| LAIG | SEM2 | 39.293km | G | 62 | 38 | - | 61.3% |
| BSCN | VAUD | 40.371km | G | 60 | 30 | - | 50.0% |
| AUT2 | FRGN | 45.057km | G | 54 | 28 | - | 51.9% |
| STEL | CCSL | 45.265km | G | 60 | 28 | - | 46.7% |
| FEUR | STF2 | 45.482km | G | 56 | 24 | - | 42.9% |
| CYLM | DOCX | 46.260km | G | 82 | 32 | - | 39.0% |
| CULA | SNNT | 48.088km | G | 62 | 32 | - | 51.6% |
| DOCX | FRGN | 49.506km | G | 62 | 26 | - | 41.9% |
| MONS | STEL | 50.483km | G | 50 | 30 | - | 60.0% |
| ALU2 | AUTN | 51.308km | G | 60 | 26 | - | 43.3% |
| CLMT | LMDM | 51.677km | G | 56 | 28 | - | 50.0% |
| STF2 | VERN | 52.434km | G | 50 | 22 | - | 44.0% |
| CLMT | STF2 | 53.075km | G | 60 | 28 | - | 46.7% |
| CYLM | PAMO | 54.582km | G | 76 | 36 | - | 47.4% |
| BRMF | FEUR | 55.210km | G | 68 | 24 | - | 35.3% |
| VAUD | VRCT | 55.899km | G | 60 | 32 | - | 53.3% |
| GDIJ | SEM2 | 56.294km | G | 56 | 32 | - | 57.1% |
| LMDM | PAMO | 56.496km | G | 48 | 30 | - | 62.5% |
| CLFD | SNNT | 67.128km | G | 68 | 30 | - | 44.1% |
| CHE2 | GRIG | 4.064km | G | 48 | 32 | 15 | 97.917% |
| EXSG | SBGS | 7.618km | G | 46 | 32 | 11 | 93.478% |
| ARNA | MAN2 | 8.242km | G | 58 | 28 | 20 | 82.759% |
| EXSG | TGRI | 10.683km | G | 48 | 30 | 10 | 83.333% |
| IGNP | MLVL | 11.918km | G | 44 | 30 | 8 | 86.364% |
| IGNP | SBGS | 20.252km | G | 46 | 30 | - | 65.2% |
| IGNP | SIRT | 21.675km | G | 44 | 30 | - | 68.2% |
| GRIG | MLVL | 26.484km | G | 48 | 32 | - | 66.7% |
| CCSL | GIE1 | 29.982km | G | 58 | 32 | - | 55.2% |
| ASNE | GIE1 | 32.876km | G | 42 | 32 | - | 76.2% |
| DHUI | SIRT | 34.866km | G | 62 | 36 | - | 58.1% |
| BAUG | NGER | 35.377km | G | 54 | 36 | - | 66.7% |
| GRIG | LIZ2 | 37.339km | G | 52 | 32 | - | 61.5% |
| AMIY | GIE1 | 39.345km | G | 52 | 26 | - | 50.0% |
| RGNC | SAUN | 40.431km | G | 62 | 36 | - | 58.1% |
| BAUG | PTRC | 41.645km | G | 48 | 26 | - | 54.2% |
| PTRC | RGNC | 46.048km | G | 56 | 24 | - | 42.9% |
| ARNA | BAUG | 49.854km | G | 62 | 30 | - | 48.4% |
| AMIY | MAGR | 50.692km | G | 54 | 36 | - | 66.7% |
| ARDN | CH2T | 51.214km | G | 54 | 28 | - | 51.9% |
| ARDN | ASNE | 51.401km | G | 46 | 28 | - | 60.9% |
| MAGR | TRYS | 52.579km | G | 70 | 34 | - | 48.6% |
| STMA | VFCH | 52.688km | G | 52 | 34 | - | 65.4% |
| TGRI | TRBL | 54.291km | G | 64 | 30 | - | 46.9% |
| ASNE | VFCH | 54.793km | G | 46 | 30 | - | 65.2% |
| CCSL | SDDP | 55.171km | G | 48 | 32 | - | 66.7% |
| CH2T | TRBL | 56.207km | G | 70 | 38 | - | 54.3% |
| MAGR | VIN1 | 58.137km | G | 50 | 28 | - | 56.0% |
| MAUP | TRBL | 59.473km | G | 86 | 42 | - | 48.8% |
| CH2T | SAUN | 61.022km | G | 50 | 32 | - | 64.0% |
| LAHE | LHVE | 2.845km | G | 66 | 28 | 33 | 92.424% |
| STJF | TANC | 4.151km | G | 50 | 30 | 19 | 98.000% |
| 1MEL | LIL2 | 4.520km | G | 50 | 32 | 14 | 92.000% |
| DGLG | DUNQ | 6.246km | G | 54 | 34 | 16 | 92.593% |
| FRAN | HONF | 6.806km | G | 52 | 34 | 16 | 96.154% |
| FRAN | LHVE | 8.449km | G | 62 | 38 | 18 | 90.323% |
| CDBC | STJF | 15.516km | G | 50 | 32 | 8 | 80.000% |
| CDBC | MOFA | 16.842km | G | 58 | 34 | 16 | 86.207% |
| HONF | TANC | 17.017km | G | 52 | 34 | 11 | 86.538% |
| CDBC | CNTL | 24.616km | G | 56 | 32 | - | 57.1% |
| HOUL | LAHE | 24.992km | G | 54 | 32 | - | 59.3% |
| HOUL | LCST | 29.656km | G | 68 | 36 | - | 52.9% |
| CNTL | VEN2 | 30.436km | G | 56 | 36 | - | 64.3% |
| CNTL | MOMO | 35.947km | G | 42 | 30 | - | 71.4% |
| CHBL | VEN2 | 41.881km | G | 46 | 34 | - | 73.9% |
| MOMO | STAB | 42.196km | G | 48 | 32 | - | 66.7% |
| BEUG | BUSI | 43.696km | G | 62 | 40 | - | 64.5% |
| BOVE | VNTE | 47.927km | G | 56 | 28 | - | 50.0% |
| AMNS | BOVE | 48.481km | G | 50 | 28 | - | 56.0% |
| VNTE | LIZ2 | 49.297km | G | 60 | 30 | - | 50.0% |
| BOVE | MOMO | 50.643km | G | 52 | 26 | - | 50.0% |
| BEUG | BVOI | 52.722km | G | 56 | 34 | - | 60.7% |
| AMNS | BEUG | 54.855km | G | 58 | 36 | - | 62.1% |
| BEUG | LIL2 | 56.809km | G | 54 | 32 | - | 59.3% |
| BVOI | EQH2 | 58.827km | G | 42 | 40 | - | 95.2% |
| DGLG | EQH2 | 64.901km | G | 46 | 34 | - | 73.9% |
| DUNQ | VLIS | 96.504km | G | 42 | 34 | - | 81.0% |
| EQH2 | SHOE | 110.236km | G | 46 | 36 | - | 78.3% |
| CHIO | SHOE | 164.136km | G | 48 | 34 | - | 70.8% |
| DIPP | STAB | 4.828km | G | 56 | 38 | 16 | 96.429% |
| BRST | GUIP | 9.498km | G | 80 | 46 | 28 | 92.500% |
| GUIP | PLDN | 10.551km | G | 80 | 42 | 32 | 92.500% |
| HERB | STNZ | 21.046km | G | 42 | 34 | - | 81.0% |
| PLST | STBR | 25.410km | G | 84 | 48 | - | 57.1% |
| NMTR | STNZ | 29.144km | G | 40 | 36 | - | 90.0% |
| LHGE | VLG2 | 29.295km | G | 64 | 48 | - | 75.0% |
| HERB | SAMR | 33.737km | G | 66 | 34 | - | 51.5% |
| PRR2 | VLG2 | 36.100km | G | 82 | 42 | - | 51.2% |
| HERB | VNNS | 38.269km | G | 48 | 40 | - | 83.3% |
| LOYT | PNTV | 41.584km | G | 92 | 38 | - | 41.3% |
| PLDN | PLGN | 42.271km | G | 48 | 34 | - | 70.8% |
| PRR2 | VLPL | 42.540km | G | 70 | 30 | - | 42.9% |
| KEGA | PLGN | 42.572km | G | 56 | 30 | - | 53.6% |
| KEGA | PNTV | 42.660km | G | 68 | 28 | - | 41.2% |
| LOYT | SAMR | 42.721km | G | 80 | 36 | - | 45.0% |
| PLMG | STBR | 44.216km | G | 78 | 44 | - | 56.4% |
| NMCU | STNZ | 46.116km | G | 50 | 34 | - | 68.0% |
| LOYT | PLST | 46.604km | G | 72 | 40 | - | 55.6% |
| BAPI | VLPL | 47.003km | G | 66 | 30 | - | 45.5% |
| NOYL | OMBA | 47.074km | G | 54 | 36 | - | 66.7% |
| LAVL | OMBA | 50.711km | G | 68 | 32 | - | 47.1% |
| BAPI | NOYL | 51.814km | G | 54 | 28 | - | 51.9% |
| DOMP | VLPL | 54.259km | G | 98 | 48 | - | 49.0% |
| BAPI | PLST | 55.214km | G | 68 | 34 | - | 50.0% |
| NGER | OMBA | 56.638km | G | 50 | 34 | - | 68.0% |
| CAMO | SCIL | 77.137km | G | 42 | 34 | - | 81.0% |
| CAMO | PLDN | 207.505km | G | 48 | 32 | - | 66.7% |
| LBTM | PLST | 8.071km | G | 68 | 36 | 1 | 54.412% |
| CHTG | LHGE | 13.824km | G | 48 | 28 | 14 | 87.500% |
| LGES | LMGS | 5.750km | G | 48 | 30 | 14 | 91.667% |
| BXME | CUBX | 9.620km | G | 56 | 34 | 17 | 91.071% |
| AUNI | SPIO | 32.694km | G | 54 | 30 | - | 55.6% |
| EGLT | UZER | 38.124km | G | 42 | 30 | - | 71.4% |
| BRG9 | MATC | 38.430km | G | 48 | 30 | - | 62.5% |
| MAZE | QUYC | 38.878km | G | 44 | 32 | - | 72.7% |
| BRE2 | CHOL | 40.714km | G | 70 | 32 | - | 45.7% |
| C2CN | GARD | 40.837km | G | 52 | 32 | - | 61.5% |
| BRG9 | LGES | 43.739km | G | 52 | 32 | - | 61.5% |
| CHOL | NGER | 49.914km | G | 60 | 28 | - | 46.7% |
| LGES | UZER | 51.730km | G | 46 | 28 | - | 60.9% |
| LCVA | MPRV | 51.946km | G | 76 | 26 | - | 34.2% |
| BARD | BRTM | 52.144km | G | 60 | 32 | - | 53.3% |
| BARD | LCVA | 54.061km | G | 64 | 30 | - | 46.9% |
| BARD | MAZE | 54.515km | G | 50 | 30 | - | 60.0% |
| AICI | SCOA | 54.520km | G | 68 | 28 | - | 41.2% |
| COCR | NVPT | 56.547km | G | 56 | 30 | - | 53.6% |
| BARD | GARD | 57.508km | G | 60 | 32 | - | 53.3% |
| COCR | MATC | 58.827km | G | 50 | 32 | - | 64.0% |
| BRE2 | NVPT | 59.156km | G | 70 | 30 | - | 42.9% |
| BXME | CAPT | 59.866km | G | 56 | 32 | - | 57.1% |
| MPRV | NVPT | 59.878km | G | 62 | 14 | - | 22.6% |
| CHOL | RYO2 | 60.438km | G | 74 | 38 | - | 51.4% |
| AICI | STS2 | 61.028km | G | 68 | 38 | - | 55.9% |
| CAPT | STS2 | 62.806km | G | 54 | 34 | - | 63.0% |
| CUBX | QUYC | 63.088km | G | 44 | 32 | - | 72.7% |
| EGLT | SMDP | 64.648km | G | 50 | 28 | - | 56.0% |
| AUNI | MAZE | 65.308km | G | 58 | 36 | - | 62.1% |
| ILDX | SPIO | 12.169km | G | 52 | 38 | 12 | 96.154% |
| TLSE | TLSG | 1.266km | G | 52 | 32 | 14 | 88.462% |
| AGDE | AGDS | 1.918km | G | 60 | 30 | 22 | 86.667% |
| CSTO | TLSG | 2.719km | G | 66 | 34 | 28 | 93.939% |
| FJC2 | LEZI | 17.578km | G | 66 | 32 | 19 | 77.273% |
| AGDS | SET1 | 20.829km | G | 60 | 28 | - | 46.7% |
| LEZI | PARD | 25.153km | G | 62 | 30 | - | 48.4% |
| CRAL | STGS | 26.785km | G | 70 | 40 | - | 57.1% |
| LACA | PARD | 28.808km | G | 56 | 26 | - | 46.4% |
| HOLA | SAFQ | 28.849km | G | 52 | 28 | - | 53.8% |
| LACA | SAFQ | 32.780km | G | 62 | 34 | - | 54.8% |
| MTIL | PRAD | 35.627km | G | 56 | 32 | - | 57.1% |
| AGN2 | C2CN | 41.560km | G | 52 | 32 | - | 61.5% |
| AUBT | PEY2 | 43.219km | G | 48 | 30 | - | 62.5% |
| CORZ | CRAL | 49.737km | G | 56 | 32 | - | 57.1% |
| CABN | LACA | 50.197km | G | 50 | 30 | - | 60.0% |
| ESCO | STGS | 51.626km | G | 66 | 40 | - | 60.6% |
| AGDE | PARD | 54.237km | G | 52 | 28 | - | 53.8% |
| CSTO | FAJP | 55.529km | G | 58 | 28 | - | 48.3% |
| CRAL | PEY2 | 57.213km | G | 58 | 40 | - | 69.0% |
| CABN | MTIL | 58.558km | G | 58 | 32 | - | 55.2% |
| AGN2 | AUBT | 59.030km | G | 44 | 28 | - | 63.6% |
| AUBT | TLSE | 59.803km | G | 54 | 28 | - | 51.9% |
| FAJP | LEZI | 62.130km | G | 72 | 26 | - | 36.1% |
| FILF | FJC2 | 62.403km | G | 50 | 28 | - | 56.0% |
| CREU | FILF | 78.654km | G | 42 | 24 | - | 57.1% |
| JOUS | SAFQ | 109.140km | G | 70 | 32 | - | 45.7% |
| BELL | EBRE | 115.284km | G | 60 | 30 | - | 50.0% |
| BELL | ESCO | 126.542km | G | 58 | 30 | - | 51.7% |
| TLMF | TLSE | 8.686km | G | 76 | 30 | 32 | 81.579% |




Fixation des ambiguités Core
| Station 1 | Station 2 | Longueur | Constellation | Ambiguités posées | Ambiguités fixées QIF | Ambiguités fixées L12 | Ratio |
|---|---|---|---|---|---|---|---|
| GRAS | NICA | 25.389km | G | 48 | 28 | - | 58.3% |
| MLVL | TGRI | 46.866km | G | 48 | 34 | - | 70.8% |
| TGRI | TRBL | 54.291km | G | 64 | 30 | - | 46.9% |
| CNTL | FRAN | 62.349km | G | 48 | 30 | - | 62.5% |
| DGLG | LIL2 | 69.994km | G | 58 | 38 | - | 65.5% |
| EGLT | LGES | 78.902km | G | 52 | 24 | - | 46.2% |
| DBMH | THNV | 85.713km | G | 48 | 34 | - | 70.8% |
| CLFD | EGLT | 91.721km | G | 56 | 28 | - | 50.0% |
| MLVL | ROMY | 94.021km | G | 48 | 30 | - | 62.5% |
| DBMH | ENTZ | 94.338km | G | 52 | 30 | - | 57.7% |
| CNTL | TGRI | 94.831km | G | 46 | 32 | - | 69.6% |
| ROMY | TRYS | 99.431km | G | 76 | 30 | - | 39.5% |
| CLFD | PUYV | 99.856km | G | 66 | 26 | - | 39.4% |
| CRAL | TLSE | 102.285km | G | 72 | 30 | - | 41.7% |
| MAN2 | TRBL | 106.913km | G | 70 | 36 | - | 51.4% |
| GAPC | GRAS | 108.635km | G | 56 | 28 | - | 50.0% |
| BRMF | PUYV | 112.426km | G | 56 | 30 | - | 53.6% |
| AXPV | GAPC | 125.818km | G | 52 | 30 | - | 57.7% |
| BRST | STBR | 129.731km | G | 84 | 52 | - | 61.9% |
| HOLA | PUYV | 130.287km | G | 54 | 26 | - | 48.1% |
| AUTN | BSCN | 133.134km | G | 60 | 30 | - | 50.0% |
| BRE2 | MAN2 | 139.398km | G | 72 | 30 | - | 41.7% |
| MAN2 | VFCH | 142.465km | G | 62 | 28 | - | 45.2% |
| AUTN | BRMF | 145.348km | G | 64 | 26 | - | 40.6% |
| HOLA | TLSE | 147.352km | G | 52 | 26 | - | 50.0% |
| AUTN | TRYS | 150.482km | G | 68 | 30 | - | 44.1% |
| BSCN | DBMH | 153.456km | G | 62 | 32 | - | 51.6% |
| BRMF | GAPC | 161.582km | G | 68 | 26 | - | 38.2% |
| LIL2 | ROMY | 166.466km | G | 48 | 32 | - | 66.7% |
| CRAL | SCOA | 168.978km | G | 48 | 30 | - | 62.5% |
| CUBX | LGES | 175.940km | G | 50 | 28 | - | 56.0% |
| MAN2 | STBR | 222.650km | G | 82 | 44 | - | 53.7% |




Estimation des ZTD finaux (coordonnées fixées) - ADDNEQ2
| Statistiques | |
|---|---|
| Paramètres explicites | 7306 |
| Paramètres implicites | 3022 |
| Paramètres ajustés | 10328 |
| Observations | 244814 |
| Degré de liberté | 234486 |
| Chi**2/DOF | 4.03 |
| Stations | 0 |
| Fichier | Début | Fin | Obs. | Par. | Dof |
|---|---|---|---|---|---|
| MET_25181O_1.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 26173 | 2007 | 24166 |
| MET_25181O_2.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 25435 | 2003 | 23432 |
| MET_25181O_3.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 26548 | 2088 | 24460 |
| MET_25181O_4.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 27441 | 1988 | 25453 |
| MET_25181O_5.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 27314 | 1905 | 25409 |
| MET_25181O_6.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 25219 | 2003 | 23216 |
| MET_25181O_7.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 26393 | 1992 | 24401 |
| MET_25181O_8.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 25775 | 1975 | 23800 |
| MET_25181O_9.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 28863 | 2241 | 26622 |




Estimation des coordonnées et des ZTD finaux - ADDNEQ2
| Statistiques | |
|---|---|
| Paramètres explicites | 8014 |
| Paramètres implicites | 3022 |
| Paramètres ajustés | 11036 |
| Observations | 244814 |
| Degré de liberté | 233778 |
| Chi**2/DOF | 3.57 |
| Stations | 236 |
 Statistiques
Statistiques Equations normales
Equations normales| Fichier | Début | Fin | Obs. | Par. | Dof |
|---|---|---|---|---|---|
| CRD_25181O_1.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 26173 | 2097 | 24076 |
| CRD_25181O_2.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 25435 | 2093 | 23342 |
| CRD_25181O_3.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 26548 | 2181 | 24367 |
| CRD_25181O_4.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 27441 | 2081 | 25360 |
| CRD_25181O_5.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 27314 | 1998 | 25316 |
| CRD_25181O_6.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 25219 | 2093 | 23126 |
| CRD_25181O_7.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 26393 | 2082 | 24311 |
| CRD_25181O_8.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 25775 | 2065 | 23710 |
| CRD_25181O_9.NQ0 | 2025-06-30 12:00:00 | 2025-07-01 00:00:00 | 28863 | 2340 | 26523 |




Mise en référence IGS14
Paramètres de transformation| Valeur | Ecart-type | |
|---|---|---|
| TX | -13.66 | - 15.79 |
| TY | 100.72 | - 20.44 |
| TZ | 44.75 | - 15.52 |
| k | -0.00098 | - 0.002 |
| RX | - 0 0 0.000840 | - 0.0005 |
| RY | - 0 0 0.002039 | - 0.0005 |
| RZ | 0 0 0.001354 | - 0.0005 |
Résidus
| Station | N (mm ) | E (mm ) | U (mm ) | Mark |
|---|---|---|---|---|
| 1MEL | 1.84 | 1.67 | 3.19 | M |
| AAER | 0.84 | -3.09 | -8.41 | |
| AGDE | 1.29 | 1.17 | -1.12 | |
| AGDS | -0.58 | 0.17 | 6.25 | |
| AGN2 | 2.76 | -2.96 | -0.57 | M |
| AICI | -8.41 | 0.69 | 1.24 | |
| ALGY | 0.78 | -4.38 | 33.09 | M |
| ALU2 | 2.24 | 0.63 | 4.02 | M |
| AMIY | -0.74 | 0.67 | -32.14 | M |
| AMNS | -2.83 | 2.01 | -2.78 | |
| ANDE | -7.82 | -9.04 | 9.67 | |
| APT2 | 11.66 | -3.04 | 30.65 | M |
| ARDN | -1.63 | -5.06 | 1.45 | |
| ARNA | 13.72 | 3.78 | 5.06 | M |
| ASNE | 2.83 | 0.04 | 5.10 | M |
| ATH2 | 0.02 | 0.35 | -1.08 | M |
| AUBT | 8.95 | 5.62 | 12.87 | M |
| AUNI | 5.03 | -1.33 | 3.56 | |
| AUT2 | 5.75 | 6.39 | 4.51 | M |
| AUTN | -5.04 | 1.03 | 6.36 | |
| AXPV | 1.66 | 0.81 | -8.42 | |
| BADH | 2.37 | -3.00 | -24.75 | |
| BAL2 | 3.62 | 3.61 | -6.31 | M |
| BANN | 3.67 | -0.27 | -13.17 | |
| BAPI | -0.77 | -1.56 | 9.04 | M |
| BARD | 5.58 | -5.91 | -2.31 | M |
| BAUG | 4.09 | -7.66 | -17.66 | M |
| BELL | 0.40 | -0.28 | -3.42 | |
| BEUG | -9.48 | -6.02 | 9.24 | M |
| BLVR | 7.34 | 1.04 | -31.42 | M |
| BOVE | -2.92 | 15.04 | -0.28 | M |
| BRE2 | 6.44 | -2.07 | 3.25 | |
| BRG9 | 13.18 | 7.18 | -1.34 | M |
| BRM2 | 3.87 | 4.08 | -0.90 | M |
| BRMF | 8.89 | -0.20 | -13.40 | M |
| BRST | -2.45 | -0.31 | -0.42 | |
| BRTM | 5.49 | -1.78 | 12.38 | M |
| BSCN | 4.44 | 2.34 | 1.18 | |
| BUSI | -2.27 | 5.84 | 4.18 | M |
| BVOI | 6.56 | 18.18 | 4.99 | M |
| BVS2 | 2.73 | -0.61 | 14.46 | M |
| BXME | 3.45 | 2.11 | -10.45 | M |
| C2CN | 10.49 | -2.26 | 18.80 | M |
| CABN | -8.56 | 0.79 | 11.48 | M |
| CAMO | -1.49 | 3.73 | 4.29 | |
| CANE | 6.30 | 4.81 | -15.94 | M |
| CAPT | -3.22 | -2.84 | 13.91 | |
| CCSL | 0.94 | -4.84 | -12.86 | M |
| CDBC | 1.46 | 3.79 | -9.81 | M |
| CERN | 1.64 | -3.59 | -16.38 | |
| CH2T | -0.10 | 13.01 | -6.98 | M |
| CHBL | -5.08 | -6.89 | -2.21 | M |
| CHE2 | 2.47 | 3.43 | 16.22 | M |
| CHIO | 0.00 | 4.07 | 2.80 | |
| CHOL | -2.58 | 3.64 | 1.78 | M |
| CHPO | -0.99 | -0.62 | -0.36 | M |
| CHTG | -6.97 | 6.07 | -37.50 | M |
| CLFD | -1.12 | 1.90 | 21.05 | |
| CLMT | -0.20 | 11.67 | 4.48 | |
| CNNE | 1.79 | 12.34 | -23.32 | M |
| CNTL | -1.44 | 1.57 | -8.64 | M |
| COCR | 0.73 | 5.37 | 4.88 | M |
| COMO | 1.04 | -2.45 | -2.64 | |
| CONN | 10.91 | -3.84 | -14.10 | M |
| CORZ | 6.35 | -4.35 | 6.94 | M |
| CRAL | 0.74 | 0.58 | -2.12 | |
| CRAU | 2.21 | 5.24 | 1.49 | M |
| CREU | -0.58 | 0.65 | -5.36 | |
| CRTS | -3.34 | 5.30 | 31.63 | M |
| CSTO | -0.69 | -3.10 | -3.69 | |
| CUBX | -4.91 | -0.98 | -9.62 | |
| CULA | -3.28 | -1.24 | -16.62 | M |
| CYLM | 1.28 | 7.67 | 0.05 | M |
| DBMH | -3.60 | 3.22 | -18.72 | M |
| DEQU | -4.78 | 1.20 | 31.01 | M |
| DGLG | -0.70 | 1.32 | 8.25 | |
| DHUI | 1.95 | -5.57 | 4.28 | M |
| DILL | 1.42 | -0.41 | -10.74 | M |
| DIPP | -0.06 | -1.16 | 1.90 | M |
| DOCX | -4.25 | 4.05 | 7.97 | M |
| DOMP | 4.35 | 1.05 | -4.74 | M |
| DRUL | -2.17 | -2.59 | -12.23 | M |
| DUNQ | 11.20 | -0.63 | -15.29 | M |
| EBNS | -29.10 | 7.80 | -0.59 | M |
| EBRE | -3.26 | 1.93 | -1.87 | |
| EGLT | 2.94 | 8.68 | -13.09 | |
| ENTZ | 2.19 | -3.89 | 3.45 | |
| EOST | -4.11 | 7.60 | 0.14 | M |
| EQH2 | 1.07 | 1.11 | -1.65 | M |
| ESCO | 0.34 | -2.50 | -0.73 | |
| ETOI | -0.89 | 4.36 | -15.44 | M |
| EXSG | -2.51 | 4.66 | 18.61 | M |
| FAJP | 0.63 | -4.75 | 4.20 | M |
| FEUR | 5.18 | 4.39 | -9.95 | M |
| FIED | 6.59 | 4.54 | -20.45 | M |
| FILF | 4.41 | -0.29 | -37.53 | M |
| FJC2 | 13.24 | 16.92 | 6.69 | M |
| FRAN | 5.44 | -12.42 | -4.68 | M |
| FRGN | -5.76 | 0.54 | -41.17 | M |
| GAJN | 8.23 | 3.12 | -13.54 | M |
| GAPC | -3.29 | -1.76 | -5.51 | M |
| GARD | 3.83 | -7.99 | 0.55 | M |
| GDIJ | 3.32 | -0.20 | 9.81 | |
| GIE1 | 2.64 | 1.22 | -3.49 | M |
| GRAC | -0.25 | 0.50 | 6.42 | |
| GRAS | 0.74 | 0.54 | 8.95 | |
| GRIG | -4.06 | 0.36 | -1.15 | M |
| GUIP | -0.95 | 4.04 | -11.18 | |
| HERB | 3.37 | -1.65 | -19.52 | M |
| HOLA | 6.57 | -3.23 | 7.79 | M |
| HONF | -3.35 | -9.48 | 12.82 | M |
| HOUL | 3.83 | 4.89 | -3.74 | M |
| IGNP | -0.05 | 5.80 | -4.41 | M |
| ILDX | 0.18 | -0.31 | -7.29 | |
| JANU | 3.58 | -5.05 | 10.59 | M |
| JOUS | 8.02 | -3.51 | 12.63 | M |
| JOUX | 2.06 | 4.04 | -14.42 | M |
| KEGA | -1.61 | 2.80 | -3.03 | M |
| LACA | 3.16 | 0.57 | -5.94 | M |
| LAHE | 5.23 | -1.35 | -2.98 | M |
| LAIG | 0.16 | 6.09 | 21.21 | M |
| LAVL | 10.96 | -4.46 | -8.25 | M |
| LBTM | -2.99 | -1.30 | -12.44 | M |
| LCST | -2.12 | 6.73 | -12.95 | M |
| LCVA | 11.17 | 4.04 | 31.63 | M |
| LDB2 | 0.86 | 8.25 | -39.20 | M |
| LESA | -2.03 | 1.87 | 17.47 | M |
| LEZI | 7.54 | -1.21 | 5.92 | M |
| LGES | 2.16 | 1.66 | 6.84 | M |
| LHGE | -0.05 | 2.66 | -7.56 | M |
| LHVE | 3.79 | 7.40 | 2.96 | M |
| LIL2 | 3.30 | -2.81 | 5.74 | |
| LIZ2 | 6.16 | 3.39 | 14.35 | M |
| LLGV | -0.31 | -1.49 | -8.40 | M |
| LMDM | 11.46 | 3.98 | -9.74 | M |
| LMGS | 2.17 | 1.73 | -13.64 | M |
| LOYT | -2.19 | 0.45 | 7.85 | M |
| LRTZ | 1.09 | 3.91 | -3.09 | |
| MAGR | 2.49 | 4.44 | -16.52 | M |
| MAN2 | 0.37 | 3.27 | -4.21 | |
| MARS | 14.02 | 4.04 | -52.96 | * |
| MATC | -0.63 | 6.89 | -12.15 | M |
| MAUP | 5.54 | -2.09 | -21.09 | M |
| MAZE | 10.17 | 1.78 | -2.99 | M |
| MEDI | -2.14 | -0.69 | 3.39 | |
| MEND | -0.88 | 2.86 | -1.55 | M |
| MICH | -5.66 | 5.87 | -7.65 | |
| MIRE | -6.73 | 0.08 | 5.70 | M |
| MLVL | -2.85 | 8.07 | -29.88 | |
| MNBL | -5.22 | 7.84 | -19.34 | M |
| MOFA | 3.20 | 5.09 | -15.46 | M |
| MOMO | -1.92 | -5.43 | 0.54 | M |
| MONS | -0.51 | -0.51 | 7.05 | M |
| MPRV | 22.98 | -10.51 | 6.81 | M |
| MTIL | -0.96 | 4.80 | 23.11 | M |
| MTLC | -1.73 | 0.82 | -1.06 | M |
| NGER | 2.87 | 3.46 | -13.65 | M |
| NICA | 3.37 | 1.93 | -0.27 | |
| NMCU | -6.79 | 0.98 | -14.95 | M |
| NMTR | -5.58 | 1.28 | -1.23 | M |
| NOGT | 5.13 | -0.93 | -12.94 | M |
| NOYL | 3.95 | 4.69 | 2.68 | M |
| NVPT | 0.51 | 3.39 | 3.34 | |
| OMBA | 2.35 | -0.17 | -1.28 | M |
| OPME | -8.35 | -28.22 | -19.70 | M |
| PAMO | 0.38 | 5.49 | -4.92 | M |
| PARD | 2.07 | -5.70 | 10.90 | M |
| PAYE | 1.99 | 4.42 | -36.10 | M |
| PEY2 | -1.05 | -13.01 | 15.01 | M |
| PLDN | 2.33 | 6.31 | 3.54 | M |
| PLGN | -2.27 | 12.08 | -8.11 | M |
| PLMG | -3.19 | 1.41 | 1.68 | M |
| PLST | -8.06 | 1.35 | 6.85 | M |
| PNTV | 0.70 | 2.94 | -8.43 | M |
| PRAD | 8.28 | -8.85 | 10.20 | M |
| PRR2 | -4.37 | 2.66 | 23.07 | M |
| PTRC | -11.01 | 7.31 | 8.73 | M |
| PUYV | -2.83 | -2.28 | 27.53 | |
| QNSN | 4.91 | -2.75 | 2.17 | M |
| QUYC | 4.50 | 2.44 | 3.24 | M |
| RGNC | 4.98 | 2.00 | -10.21 | M |
| RMZT | 3.92 | -0.78 | 11.70 | M |
| ROMY | -11.12 | -4.63 | -28.05 | M |
| RUPT | 3.23 | -0.87 | 8.58 | M |
| RYO2 | -8.23 | 0.88 | 10.89 | M |
| SAFQ | 4.49 | 0.57 | 30.55 | M |
| SAMR | 1.61 | 8.73 | 0.82 | M |
| SAUN | -0.46 | 4.32 | 6.26 | |
| SBGS | 6.27 | -1.76 | 9.25 | |
| SCIL | 3.55 | 4.44 | 5.96 | |
| SCOA | -7.72 | -10.93 | -12.21 | |
| SDDP | -2.59 | 2.24 | 1.55 | M |
| SEM2 | 1.39 | -1.09 | 14.68 | M |
| SET1 | 3.62 | -0.51 | 0.22 | M |
| SHOE | -4.23 | 1.19 | 6.61 | |
| SIRT | -3.08 | 1.02 | -3.46 | M |
| SIST | -9.16 | 7.94 | 1.84 | M |
| SMCR | 2.42 | 2.81 | -18.93 | M |
| SMDP | -1.01 | -0.03 | -9.95 | M |
| SNNT | -0.70 | 4.99 | 11.28 | M |
| SPIO | 2.33 | -1.98 | 3.30 | M |
| STAB | 3.57 | -2.64 | 11.34 | M |
| STAZ | -1.86 | -3.79 | -2.14 | M |
| STBR | 3.60 | 5.70 | -18.04 | M |
| STEL | 8.46 | 9.11 | -7.46 | M |
| STF2 | 8.91 | 0.04 | -12.08 | M |
| STGS | -2.62 | -5.38 | -3.76 | M |
| STJF | 0.01 | 4.28 | -7.29 | M |
| STMA | 16.04 | -8.80 | -6.43 | |
| STMR | -3.68 | 0.93 | -0.00 | M |
| STNZ | 0.54 | 3.59 | -3.03 | M |
| STS2 | -12.86 | -5.48 | -7.40 | M |
| TANC | 3.70 | 2.53 | -2.99 | M |
| TGRI | 4.15 | 0.96 | 15.31 | M |
| THNV | 1.81 | -1.65 | -4.31 | |
| TLMF | -1.86 | -0.37 | 0.46 | |
| TLSE | -0.68 | -2.01 | 5.85 | |
| TLSG | 0.97 | -3.44 | 1.49 | |
| TLTG | 5.48 | 0.63 | -1.42 | M |
| TRBL | 0.55 | -1.11 | 3.10 | M |
| TRYS | -5.42 | 2.32 | -4.49 | |
| UZER | 2.54 | 7.81 | 3.92 | M |
| VAUD | 3.41 | -5.85 | 22.19 | |
| VEN2 | -0.77 | 0.49 | 5.27 | M |
| VERN | 7.47 | 0.44 | -22.84 | M |
| VFCH | 0.67 | -2.36 | -1.88 | |
| VIN1 | 7.44 | 3.77 | -16.70 | M |
| VLG2 | -0.38 | 0.27 | 1.05 | M |
| VLIS | 0.89 | 1.83 | -3.57 | |
| VLPL | -2.14 | 5.66 | 0.26 | M |
| VNNS | -3.37 | 2.92 | -8.67 | M |
| VNTE | 7.77 | -4.09 | 20.89 | M |
| VOUZ | -0.23 | 3.71 | 12.37 | M |
| VRCT | 11.14 | 4.00 | 13.57 | M |
| VSOU | -5.35 | 0.88 | -4.34 | M |
| YSCN | 6.03 | -0.09 | -8.79 | M |